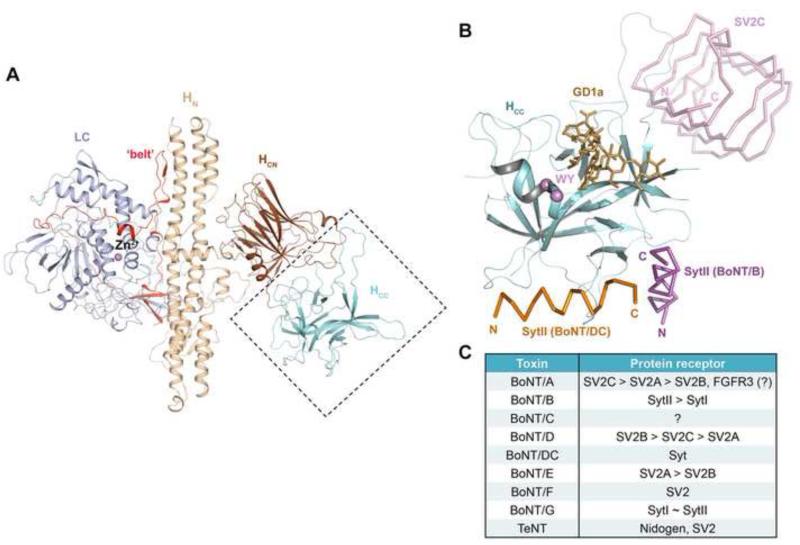
Figure 1.
The conserved structure of BoNTs and their diverse receptor-binding modes. (A) The conserved architecture of BoNTs: LC (light blue), HN (wheat), the belt region connecting LC and HN (red), HCN (brown), and HCC (cyan) (BoNT/B, PDB ID: 2NP0). (B) The HCC is a versatile receptor-binding domain. The model is built based on superposition of the structures of HCB–SytII (PDB ID: 4KBB), HCDC–SytII (PDB ID: 4ISR), and HCA–SV2C (PDB ID: 4JRA). HCCB is represented as cartoon (cyan) and the view direction is similar to that shown in panel (A). The BoNT/B-bound SytII (magenta), BoNT/DC-bound SytII (orange), and BoNT/A-bound SV2C (pink) are drawn in ribbon. GD1a, representing the polysialo-ganglioside receptor, is represented as sticks (gold). The highly conserved ganglioside-binding residues (WY) are highlighted in purple. (C) A summary of the protein receptors of various BoNT serotypes and tetanus toxin (TeNT). The preferred receptors of BoNT/A, /B, /D and /E are listed in the order of descending affinities. SytI and SytII show similar binding affinity towards BoNT/G.