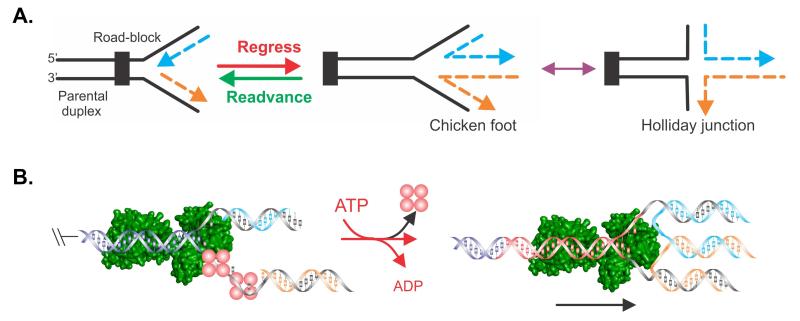
Figure 1. Schematic of events that could transpire at a stalled DNA replication fork.
(A). The fork is shown impeded and the nascent leading (blue) and lagging (orange) strands indicated. Fork regression (red arrow) would result in rightward movement of the fork, away from the site of the replication road-block, concomitant with the extrusion of a duplex region resulting from the annealing of the nascent leading and lagging strands. The resulting DNA structure has been termed the “chicken foot” and is structurally similar to a Holliday junction. Fork readvancement (not driven by RecG) would result in leftward movement of the fork in a reaction that is likely catalyzed after regression and impediment removal or repair. (B). Model of a possible mechanism of fork regression by RecG (green). Unreplicated DNA ahead of the previously advancing fork is shown in dark blue. The nascent leading and lagging strands are coloured to match those in panel A. Red spheres, SSB. Black arrow in the right panel indicates the direction of RecG translocation and consequently, of fork regression. The nascent, reannealed parental duplex is coloured black.