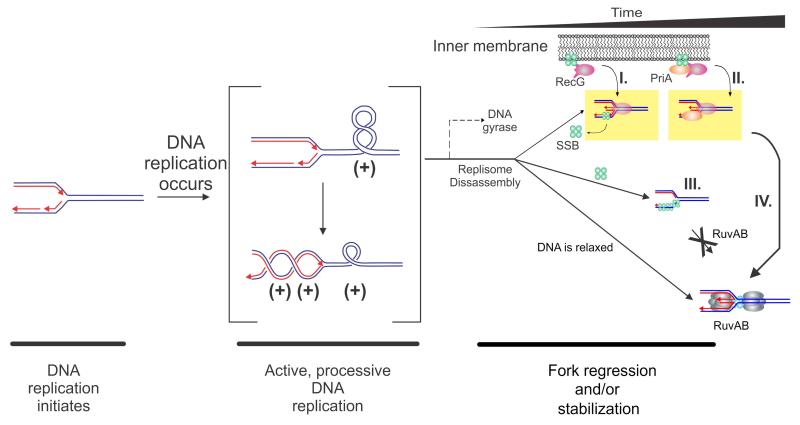
Figure 6. Mechanisms for targeting RecG to a stalled DNA replication fork.
A model of the topological domains of a segment of the E. coli chromosome undergoing replication is shown. This figure is adapted from (Buss et al., 2008; Slocum et al., 2007). Parental DNA is coloured blue and nascent daughter DNA is coloured red with arrowheads indicating 3′-ends. The RecG-SSB and RecG-PriA-SSB complexes are shown associated with the inner membrane as suggested by Yu et al (Yu et al., 2014). Once the fork encounters a block, one of several temporally spaced events may occur with RecG +/-SSB (and/or PriA) acting first followed by RuvAB. (I) If DNA gyrase acts prior to the dissociation of the replication machinery (i.e., within the 5-7 minute window following fork stalling), the (+)scDNA is converted to (-)scDNA. RecG (either in complex with SSB or released from SSB) binds to the (-)scDNA and drives fork regression. If the replisome disassembles exposing gaps in either strand, the gap will be rapidly bound by SSB (green spheres) in complex with RecG (pink), which then regresses the fork. RecG may displace SSB or co-translocation may occur, but this remains an open question. (II). The replisome disassembles and is instead bound RecG-SSB-PriA, leading to fork stabilization as suggested by Tanaka (Tanaka and Masai, 2006). (III). Following replisome disassembly, regions of exposed ssDNA are bound by SSB. SSB inhibits RuvAB, but is a target for either RecG, RecG-SSB, PriA or PriA-SSB. (IV), Once fork processing has taken place, a substrate resembling a Holliday junction is formed and is then acted upon by RuvAB