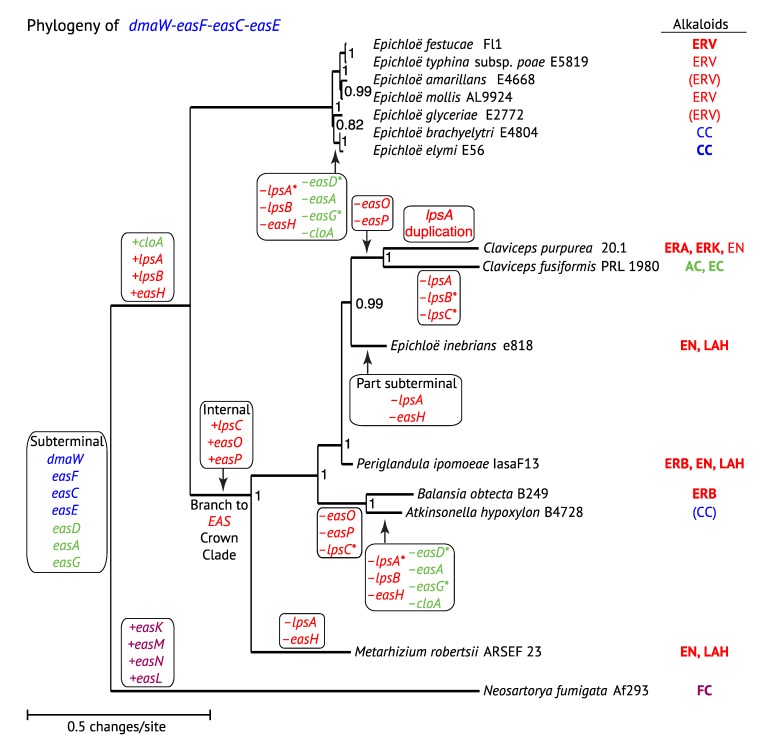
Figure 3.
Phylogeny of concatenated dmaW-easF-easC-easE genes. The phylogenetic tree is based on a nucleotide alignment of coding sequences of the core genes for the first four steps in ergot alkaloid biosynthesis available from sequenced genomes. Sequences were aligned with MUSCLE [41], and trees were inferred by maximum likelihood with PhyML implemented by Phylogeny.fr [40]. Node support was determined by the approximate likelihood ratio test [42]. Gene gains and loses are indicated by + and –, respectively, and asterisks (*) indicate that remnants or pseudogenes can be found in one or more members of the clade. Genes are color-coded based on position of the encoded step within the pathway. The major pathway end product of each strain is indicated on the right in bold face (product produced) or regular type (product predicted but not yet tested), or in parentheses (product predicted but undetected).