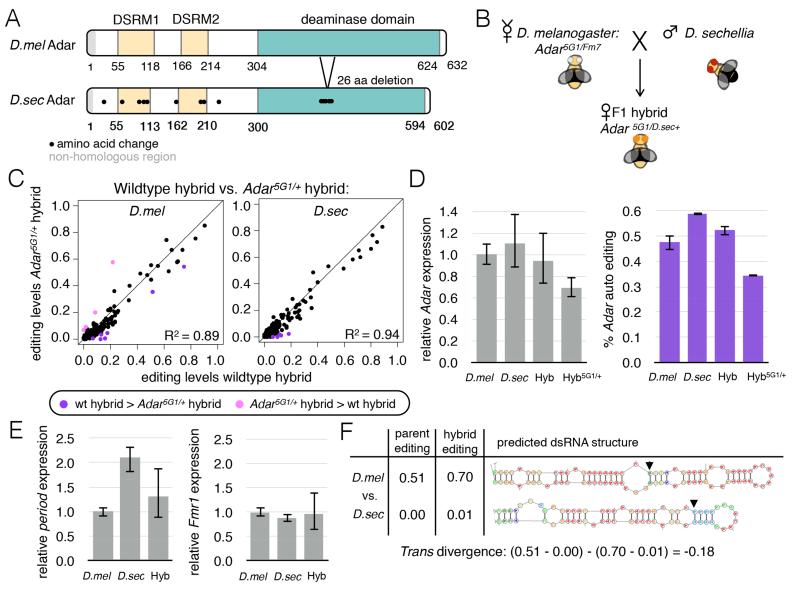
Figure 4. Differences in D. melanogaster and D. sechellia Adar proteins are not responsible for editing level differences between the species.
(A) Schematic highlighting amino acid differences between D. melanogaster and D. sechellia Adar proteins. (B) Schematic depicting the cross to create hybrid flies with only D. sechellia Adar using the Adar5G1 mutant (Palladino et al., 2000). (C) Scatter plots comparing editing levels between wildtype hybrids and Adar5G1/D.sec+ hybrids in D. melanogaster (left) and D. sechellia (right) alleles (see also Fig S3). Purple dot, wildtype hybrid more highly edited. Pink dot, Adar5G1/D.sec+ hybrid more highly edited (Fisher’s exact tests, FDR=5%). (D) Adar expression by qPCR (left) and editing level measured from Sanger sequencing at Adar auto-regulatory editing site (right) in D. melanogaster and D. sechellia parents, wildtype hybrids and Adar5G1/D.sec+ hybrids, relative to D. melanogaster parents. Error bars = SEM. (E) Expression of period and Fmr1 mRNAs in D. melanogaster, D. sechellia and F1 hybrids measured by qPCR. Error bars = SEM. (F) Editing level and secondary structure changes between species at chr3R@1062097 which showed cis and trans effects. Arrows indicate edited adenosine.