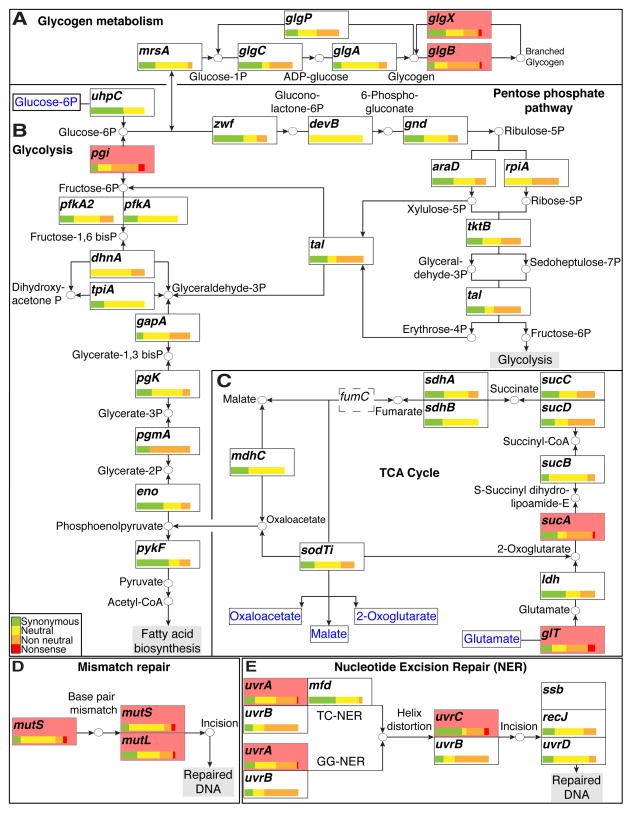
Figure 2. C. trachomatis tolerates perturbations in the tricarboxylic cycle and DNA repair networks.
(A–C) The LGV C. t central carbon metabolic network. The distribution of all synonymous (green), neutral (yellow), non-neutral (orange), and nonsense (red) bearing alleles is depicted for each ORF in the network. Distributions are normalized to total number of SNV bearing alleles identified per ORF. All ORFs with observed nonsense alleles are shaded in light red. (A) Production and break down of branched glycogen is not necessary as LGV C. t tolerates nonsense mutations in glgB and glgX. (B) The presence of a nonsense mutation in pgi suggests that the pentose phosphate pathway is sufficient to circumvent loss of fructose-6-phosphate production when the EMP pathway (Glycolysis) is blocked. (C) Nonsense mutations in glt and sucA suggest that LGV biovars can sustain metabolic activity with a severely curtailed TCA cycle. The TCA cycle is further truncated due the presence of a fumC (dashed box) pseudogene. All compounds imported from the host are shown in blue. (D) LGV C. t biovars can tolerate defects in their DNA mismatch repair machinery as suggested by the presence of nonsense bearing alleles in mutS and mutL. (E) Defects in the nucleotide excision repair pathway are also tolerated as evidenced by the presence of nonsense bearing alleles in uvrA and uvrC.