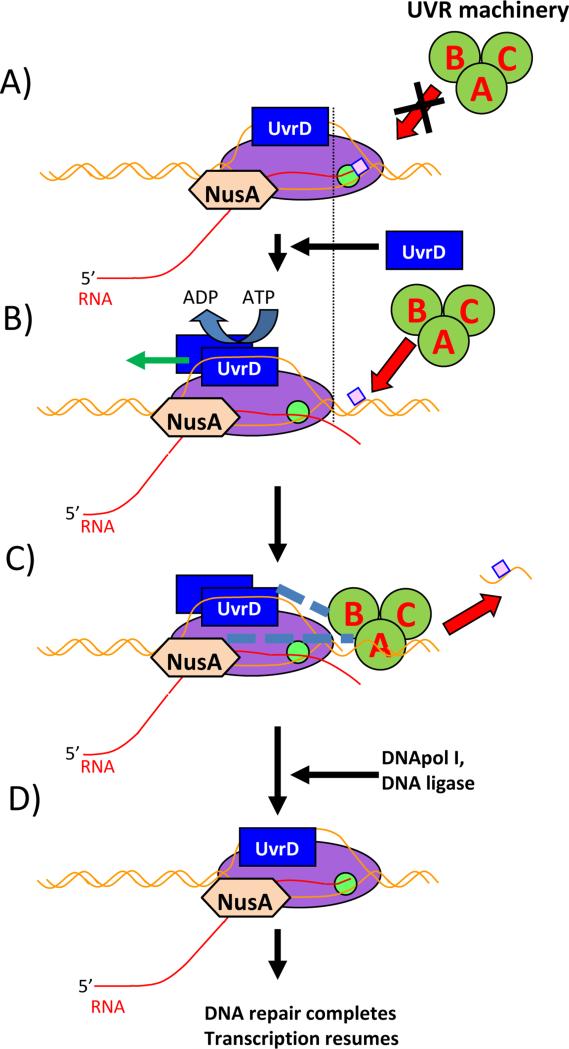
Figure 2.
Principal scheme of proposed UvrD assisted TCR in bacteria. RNAP – magenta oval, active site – green circle, UvrD – blue rectangle, UvrABC system – dark green circles, bulky lesion - pink square, NusA – light brown hexagon. DNA – brown line, RNA – red line. Proposed interactions are shown as dashed lines. A: RNAP encounters a bulky lesion on the template strand and stalls, unable to elongate. Elongation Complex blocks UvrABC from loading onto the DNA. B: UvrD forms a dimer and moves the Elongation Complex backward by using helicase/translocase activity. NusA assists UvrD, and the lesion becomes exposed. C: The UvrABC system is recruited at the DNA damage site and removes the lesion. D: UvrABC and one of the UvrD monomers leave the complex; DNApol and DNA ligase repair DNA. RNAP resumes elongation.