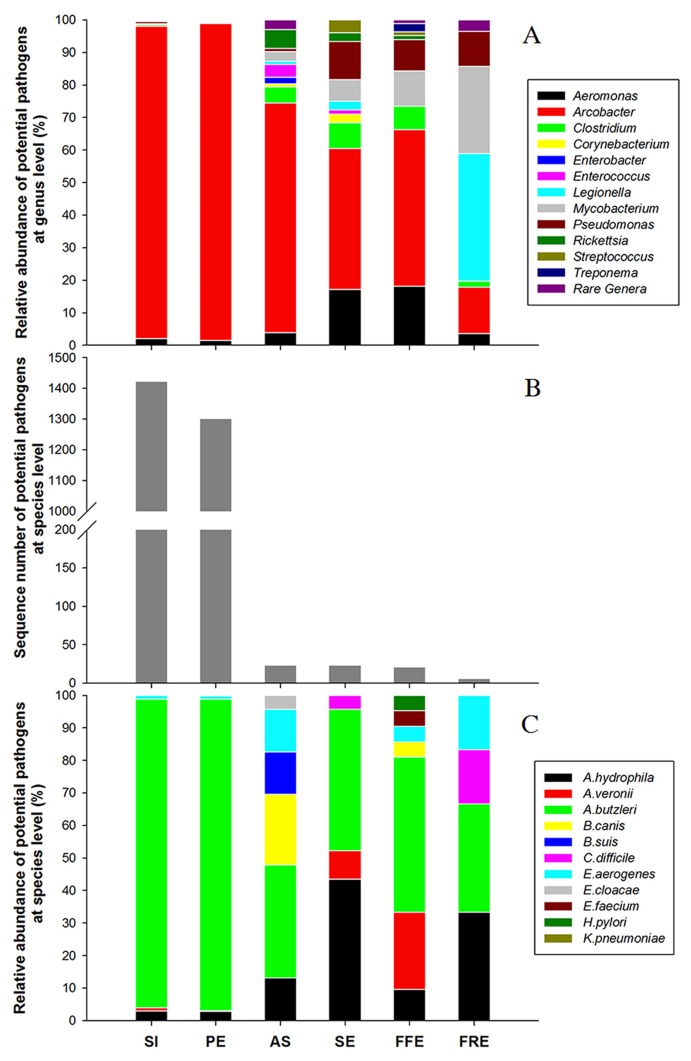
Fig 1. Occurrence and abundance of potential pathogens at genus and species levels revealed by 454 pyrosequencing of 16S rRNA gene amplicons.
(A) Relative abundance of potential pathogens at genus level. The effective pyrosequencing reads were classified using MEGAN. Rare genera refer to the taxa with maximum abundance <1% in any sample, including Bacillus, Campylobacter, Helicobacter, Klebsiella, Leptospira, Neisseria and Serratia. (B) Sequence number of potential pathogens at species level under the same sequencing depth (6200 reads). (C) Relative abundance of potential pathogens at species level. All the effective sequences identified as potentially pathogenic species were aligned using the local BLASTN tool. The relative abundance was obtained by normalizing the sequence number of each pathogenic taxon to the total number of all pathogenic taxa in one sample.