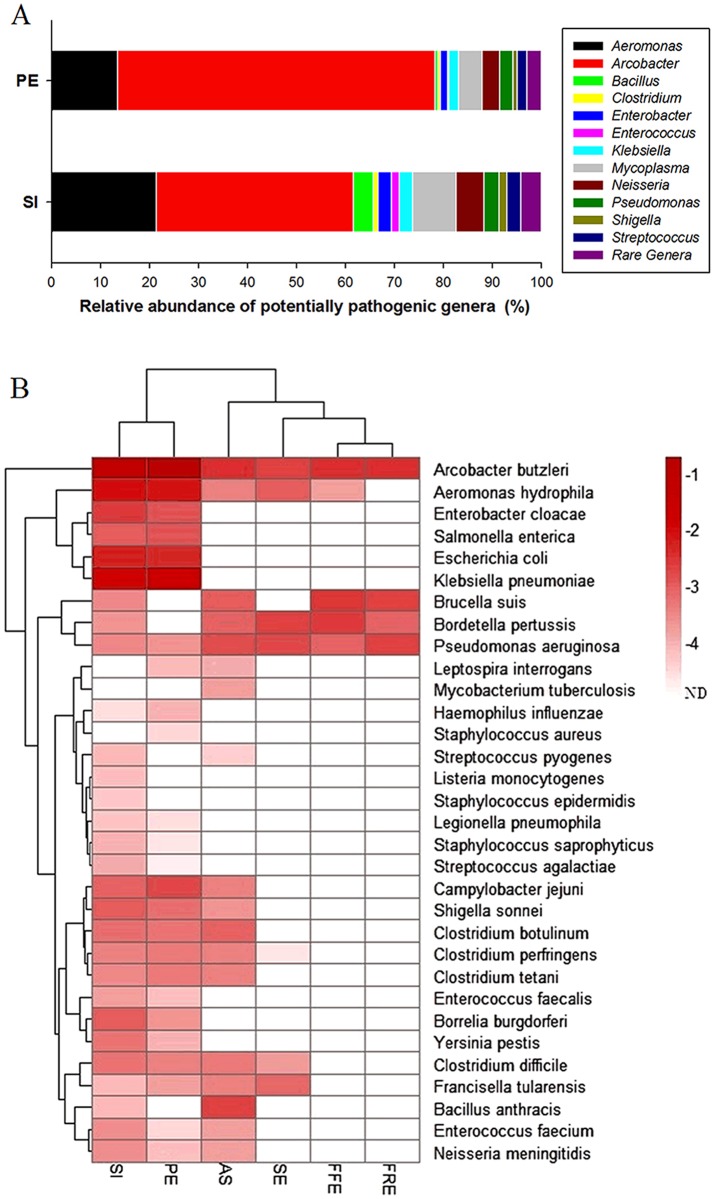
Fig 2. Diversity and relative abundance of potential pathogens at genus (A) and species (B) levels revealed by Illumina shotgun sequencing of the metagenomes.
(A) Relative abundance of potentially pathogenic genera. The denoised reads were aligned using MEGAN against 16S rRNA gene Silva database. Rare genera, indicating the taxa with maximum abundance lower than 1% in any sample, include Bordetella, Campylobacter, Corynebacterium, Escherichia, Francisella, Haemophilus, Helicobacter, Legionella, Leptospira, Listeria, Mycobacterium, Salmonella, Serratia, Staphylococcus, Treponema, Vibrio and Yersinia. (B) Heat map illustrating relative abundance (log) of potential pathogens at species level generated by MetaPhlAn. The relative abundance was obtained by normalizing the sequence number of each pathogenic taxon to the total number of all pathogenic taxa in one sample.