Fig 1. Primary structure analysis of PfalMre11.
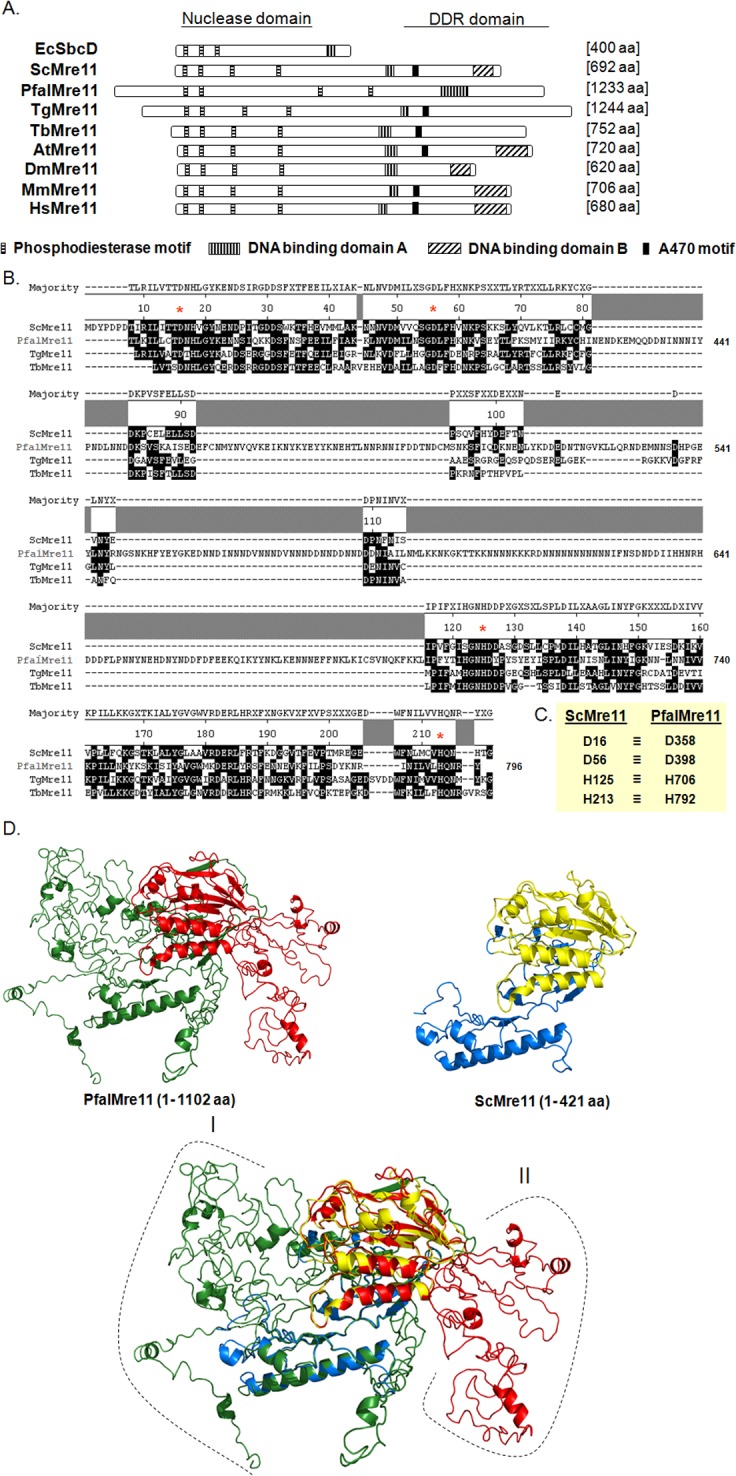
(A). Schematic representation of different domains and motifs of PfalMre11 in comparison with other Mre11 proteins from bacteria (EcSbcD); yeast (ScMre11); Toxoplasma gondii (TgMre11); Trypanosoma brucei (TbMre11); Arabidopsis thaliana (AtMre11); Drosophila melanogaster (DmMre11); mouse (MmMre11) and human (HsMre11). The four phosphodiesterase motifs within the nuclease domain are not only conserved among all the eukaryotic Mre11 but also are present in E. coli SbcD nuclease protein. PfalMre11 lacks both DNA binding domain-B and A470 motif (AV*(E/K)FV(E/D)K(D/E)(D/E)K*A, where the asterisk refers to sites that accommodate multiple residues). (B) Multiple sequence alignment showing sequence conservation within the nuclease domain of PfalMre11 with ScMre11 and other parasitic Mre11 proteins (TgMre11 and TbMre11). The functionally critical amino-acid residues in each phosphodiesterase motifs (namely, D358, D398, H706 and H792) are marked by red asterisk. The coordinates of the amino-acids positions of ScMre11 is given on the top and that of the PfalMre11 is given on the right. (C) The four critical amino-acid residues of each of the phosphodiesterase motif of ScMre11 (D16, D56, H125 and H213) and the corresponding amino-acids of PfalMre11 are shown. (D) Predicted three-dimensional structure of PfalMre11 N-terminal domain. Homology models of PfalMre11 N-terminal region (amino-acids 1–1102) and ScMre11 N-terminal region (amino-acids 1–421) are shown. The nuclease domain of PfalMre11 is shown in red, while the rest of the N-terminal is shown in green. Similarly, the nuclease domain of ScMre11 is shown in yellow, while the rest of the N-terminal is shown in marine blue. The long N-terminal extension (region I: amino-acids 1–349) and several stretches of insertions within the nuclease domain (region II) of PfalMre11 are also indicated.
