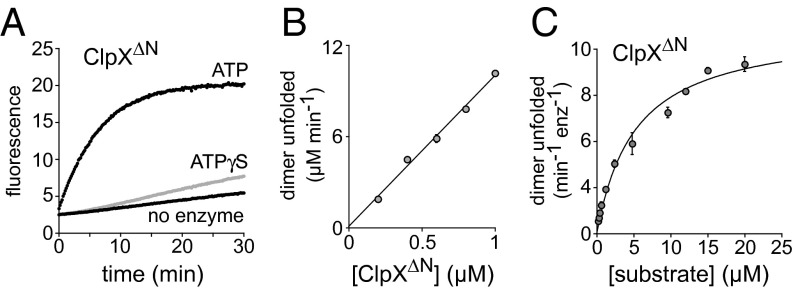
Fig. 2.
ClpX catalyzes unfolding of Arc-Gcn4-ssrA dimers. (A) Unfolding of Arc-Gcn4-ssrA by ClpX∆N was monitored by emission of acceptor fluorescence (arbitrary units). ATP supported robust unfolding relative to no enzyme, which represents spontaneous unfolding and mixing of donor- and acceptor-labeled Arc-Gcn4-ssrA dimers. ATPγS supported faster unfolding than the no-enzyme control and is hydrolyzed at ∼5% of the ATP rate (24). When present, the ClpX∆N concentration was 0.2 µM (hexamer equivalents), nucleotide concentrations were 5 mM, and donor- and acceptor-labeled Arc-Gcn4-ssrA were each present at 7.5 µM dimer equivalents. Curves are averages of two or three replicates. (B) Initial rates of Arc-Gcn4-ssrA dimer unfolding (10 µM donor labeled; 10 µM acceptor labeled) were determined using different ClpX∆N concentrations in the presence of ATP (5 mM). Data points represent averages (n = 2) ± SD. The line is a linear fit: y = 0.125 + 10x (R = 0.998). (C) Initial rates of substrate unfolding by ClpX∆N (0.2 µM) were determined for increasing concentrations of an equimolar mixture of donor-labeled and acceptor-labeled Arc-Gcn4-ssrA in the presence of ATP (5 mM). The line is a fit to the Michaelis–Menten equation (R = 0.992) with a Vmax of 11.3 ± 0.45 dimers unfolded min−1⋅enz−1 and a KM of 5.5 ± 0.7 µM. Data points are averages (n ≥ 3) ± SD.