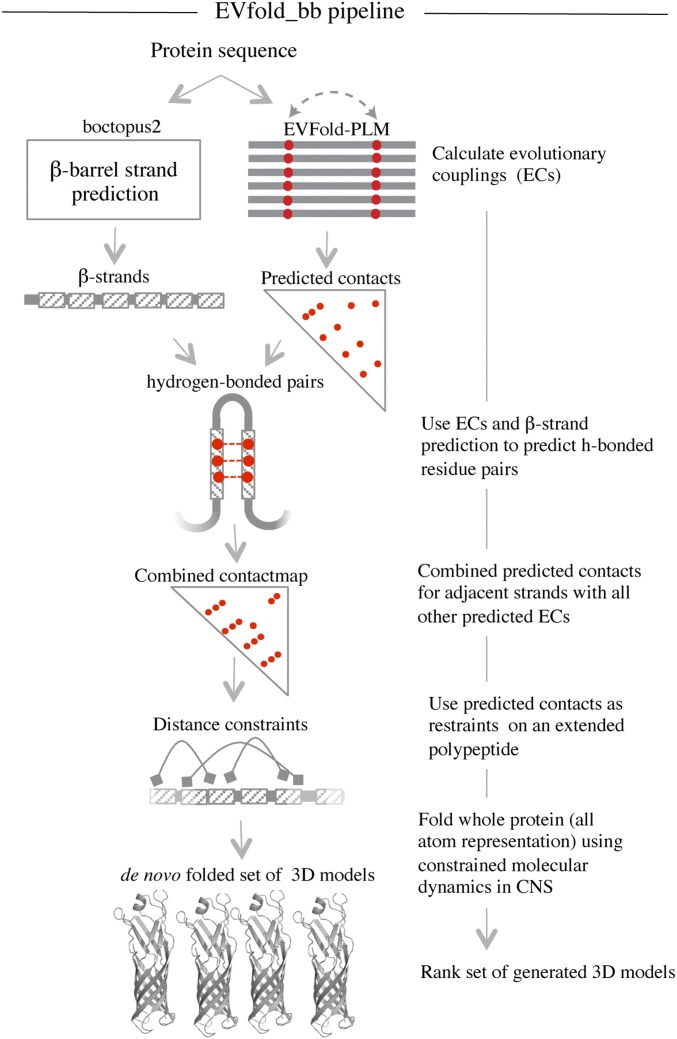
Fig. 1.
EVfold_bb pipeline to de novo fold transmembrane β-barrels. EVFold-PLM is used to generate evolutionary couplings (ECs) from a multiple-sequence alignment of the target protein. Boctopus2 is used to assign β-strands. Alternative strand registrations are compared for successive relative shifts of adjacent strands up to plus or minus three residues. The configuration with the largest sum of EC values is chosen and distance constraints are applied on N–O atoms of alternate residue pairs. In addition, other nonstrand–strand constraints are used to de novo fold the protein. Multiple models are generated and blindly ranked.