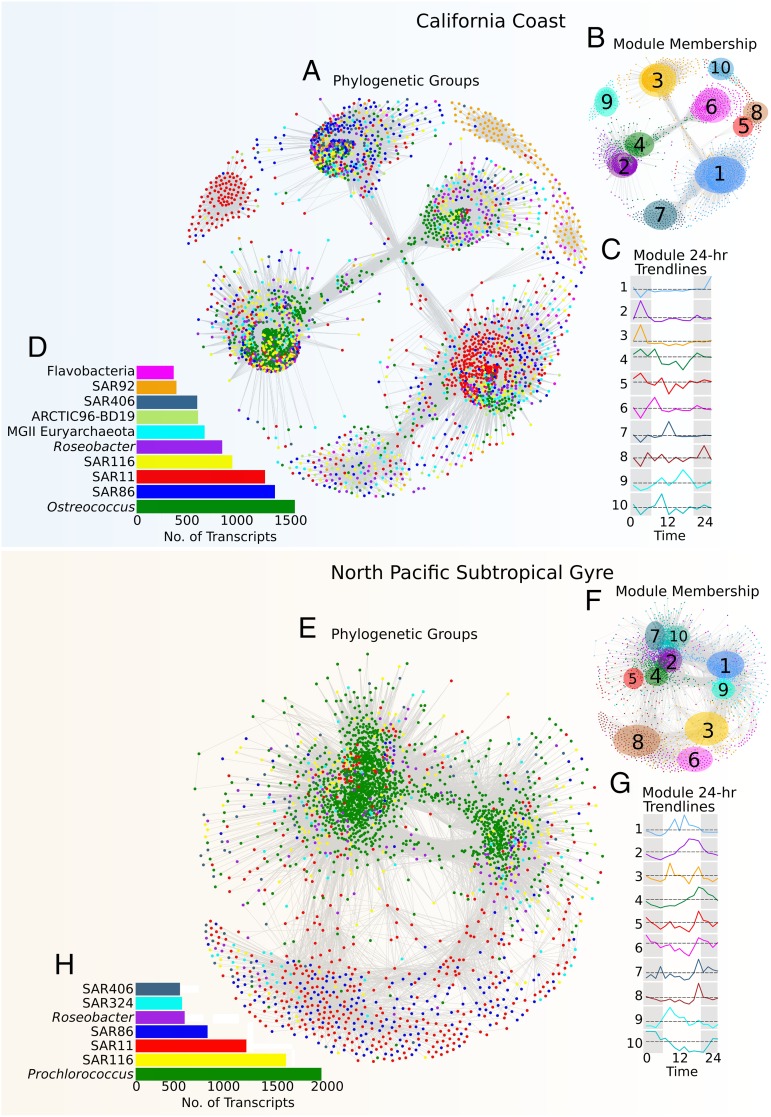
Fig. 1.
Whole-community transcriptional networks in coastal and pelagic marine microbial plankton are predominantly structured around the 24-h metabolic cycles of the photoautotrophic members. Whole-community transcriptome networks represent correlations across transcripts identified in California coast (CC) and North Pacific Subtropical Gyre (NPSG) planktonic microbial assemblages. Networks are colored according to microbial group (CC: A; NPSG: E) and module (i.e., set of coexpressed transcripts; CC: B; NPSG: F). Nodes in the network represent transcripts, whereas edges represent high pair-wise topological overlap scores between transcripts. Inset bar charts represent the total number of high abundance transcripts to which reads could be assigned in each dataset (Methods). Trendlines representing the average 24-h expression profile for the 10 largest transcriptional modules are given (CC: C; NPSG: G), with colors matching module assignments and dashed lines indicating profile averages. Both correlated and anticorrelated transcripts are present in each module, with the overall profile aligned to the predominant expression value identified. The overall number of transcripts identified in each phylogenetic group is given in the bar charts (CC: D; NPSG: H), with colors matching those shown in the main networks (A and E).