Abstract
Background and Objectives
Helicobacter pylori has been strongly associated with peptic ulcer diseases, chronic gastritis, ulcers, and reported as a risk factor for gastric cancer, too. The vaculating cytotoxin (vacA), the cytotoxin associated genes (cagA), the induced by contact with epithelium factor antigen (iceA gene), blood adhesion binding antigen (babA2), and outer membrane protein oipA have been described as different virulence factors of H. pylori. The aim of this study was to investigate the prevalence of the vacA, cagA, cagE, iceA, babA2 and oipA genotypes of H. pylori isolates from patients with upper gasterointestinal problem or dyspepsia.
Material and Methods
H. pylori isolated from endoscopic biopsies obtained from 222 studied patients. PCR was done only on cultured positive samples. The vacA alleles, cagA, cagE, iceA, babA2 and oipA genotypes were determined by PCR.
Results
The isolation rate of H. pylori strains from culture of gastric biopsies was 16.7%. The vacA alleles s1, s2, m1 and m2 were detected in 20 (54.1%), 14 (37.8%), 9 (24.3%) and 23 (62.2%) isolates, respectively. VacA s1c genotype was detected in 70.3% of isolates. s1m2 was the most frequent vacA allelic combination in the examined H. pylori strains. The cagA gene was detected in 62.2%, cagE in 40.5%, iceA1 in 48.6%, iceA2 in 16.2%, oipA in 81.1% (95% CI: 0.0902-0.1798) and babA2 in 94.6% (95% CI: 0.113- 0.207). A significant correlation was observed between vacAs1 and cagA genotypes (P<0.008), vacAs1/cagE (P=0.001), vacAs2/cagA (P<0.047), and vacAs2/cagE (P=0.016) with Non-ulcer dyspepsia; but there were not observed any correlation between other virulence markers.
Conclusion
No significant correlation was found between the existence of vacA, cagA, cagE, iceA, babA2, and oipA genes with peptic ulcer diseases and non-ulcer dyspepsia groups of studied patients.
Keywords: Helicobacter pylori, Prevalence, genotypes, gastrointestinal diseases
INTRODUCTION
Helicobacter pylori infection is one of the most common infectious diseases all over the world. It is responsible for a remarkable number of illness and abdominal pain (1). More than half of the world’s population is infected with this organism. H. pylori plays role in occurrence of gastric and duodenal cancers and intestinal lymphoma. Numerous genes such as vacA, cagA, cagE, iceA, babA and oipA, have been recognized as an important cause of pathogenesis of H. pylori infection (2-5). The cytotoxin-associated gene product (cagA), the vacuolating toxin (vacA), and the adhesion protein babA2 are major virulence factors of H. pylori that have been described (4). The severity of diseases caused by strains which express babA is greater than diseases by strains that do not express the gene. The presence of the cagE gene has also been associated with more severe clinical outcomes (5). The induced by contact with epithelium (iceA) gene has two main allelic variants iceA1 and iceA2. The expression of iceA1 is up-regulated on contact between H. pylori and human epithelial cells, and may be related with peptic ulcer disease. The expression of the outer inflammatory protein A (oipA) associated with IL-8 induction and is related with severe clinical outcomes (5). Even though H. pylori infection is common in Iran, there is only a few information about the genotyping of H. pylori strains (6,7). The genotype determination of H. pylori isolates from infected individuals with higher risk for severe diseases may lead to further knowledge about the relationship between supposed virulence genes and clinical signs. The aim of this study was to investigate the vacA, cagA, cagE, iceA, babA2, and oipA genotypes of H. pylori and their correlation with clinical diseases in patients referred to endoscopy ward of the Beheshti hospital in Kashan, Iran.
METHODS AND SUBJECTS
Study populations
Two hundred and twenty two patients with signs of abdominal pain or burning, nausea, vomiting, frequent burping, bloating and weight loss with an average age of 44.69 ± 18 years (range from 16 to 88) had undergone endoscopic investigation at Beheshti hospital in Kashan, Iran, from July 2010 through Jun 2012. H. pylori strains were isolated from the gastric mucosa biopsies specimens of H. pylori infected patients. Patients who received H. pylori eradication therapy protocol or treatment with antibiotics, bismuth-containing compounds, H2-receptor blockers, or proton pump inhibitors within 4 weeks prior to the study were excluded from the study. Informed consent was obtained from all participants, and the study was approved by the ethics committees of Kashan University of Medical Sciences.
H. pylori culture
Three gastric mucosal biopsy specimens were obtained from each patient. Specimens were used for culture, the rapid urease test, and pathological examination. One antral and one corpus specimen were directly inoculated onto the agar gel to perform the rapid urease test (RUT). The results were recorded within 24 hours. A positive RUT was indicated when the color changed from yellow to pink. The culture positive and/or positive RUTs specimens were used for chromosomal DNA extraction if the culture was negative.
Each specimen was immediately placed into Stuart’s transport medium and sent to the laboratory within 2hrs at 4°C. The biopsy specimens were smeared on the surface of Columbia agar plates supplemented with 10% horse serum and a set of antibiotics including 5 mg/l trimethoprim, 10 mg/l vancomycin, 5 mg/l cefsulodin, and 5 mg/l amphotericin B. Then plates were incubated at 37°C under microaerophilic conditions (5% O2, 10% CO2, and 85% N2), and examined after 7 days of incubation. The isolates were identified by Gram staining of the colonies, typical cell morphology, and testing for the presence of urease, oxidase, and catalase.
Chromosomal DNA extraction
The genotype profiles of H. pylori isolates were determined by PCR. Chromosomal DNA was extracted from confluent plate cultures expanded from a single colony using a commercially available kit (QIAGEN Inc., Valencia, CA, USA). Primer sequences, sizes, conditions of PCR amplifications of the glmM gene for detection and confirmation of H. pylori, the virulence genes which were designed based on published papers with a modification of PCR mixtures, and PCR conditions are summarized in Table 1. Each PCR of glmM, vacA, cagA, cagE, iceA, babA2, and oipA was performed in a total volume of 50μl containing 100ng genomic DNA from H. pylori culture, 200 μM each of dNTP, 1×PCR buffer (20 mM Tris-HCl, pH 8.4), 50 mM KCl, 1.5 mM MgCl2 (2 mM MgCl2 for CagA), 0.5μM of each primer (0.2 μM for babA2 and 0.3 μM for CagA), and 1.5 units of Taq polymerase. Negative controls were added to each PCR run including all reagents except template DNA which was substituted with ultrapure water. Aliquots of amplified samples (10 μl) were electrophoresed on 1.5-2% agarose gel in TAE buffer. The gel was stained with ethidium bromide 0.5 μg/ml. The amplified bands were visualized under ultraviolet light and photographed.
Table 1.
Primer sets used for genotyping H. pylori by PCR.
| Genes | Primer sequence (5′ à3′.) | PCR product (bp) | PCR conditions | References |
|---|---|---|---|---|
| glmM | AAGCTTTTAGGGGTGTTAGGGGTTT | 294 | 93°C, 1 min; 55°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| AAGCTTACTTTCTAACACTAACGC | ||||
| vacA s1/s2 | ATGGAAATACAACAAACACAC | 259/286 | 94°C, 1 min; 52°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| CTGCTTGAATGCGCCAAAC | ||||
| s1a | GTCAGCATCACACCGCAAC | 190 | 94°C, 1 min; 52°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| CTGCTTGAATGCGCCAAAC | ||||
| s1b | AGCGCCATACCGCAAGAG | 187 | 94°C, 1 min; 52°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| CTGCTTGAATGCGCCAAAC | ||||
| s1c | CTCTCGCTTTAGTGGGGYT | 213 | 94°C, 1 min; 52°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| CTGCTTGAATGCGCCAAAC | ||||
| m1/m2 | CAATCTGTCCAATCAAGCGAG | 567/642 | 94°C, 1 min; 52°C, 1 min; 72°C, 1 min (35 cycles) | 10 |
| GCGTCAAAATAATTCCAAGG | ||||
| cagA | ATAATGCTAAATTAGACAACTTGAGCGA | 298 | 94°C, 1 min; 60°C, 1 min; 72°C, 1 min (45 cycles) | 10 |
| TTAGAATAATCAACAAACATCACGCCAT | ||||
| cagE | TTGAAAACTTCAAGGATAGGATAGAGC | 508 | 94°C, 1 min; 53°C, 45 s; 72°C, 45 s (35 cycles) | 10 |
| GCCTAGCGTAATATCACCATTACCC | ||||
| iceA1 | GTGTTTTTAACCAAAGTATC | 247 | 95°C 1 min; 57°C, 1 s; 72°C, 1 min (35 cycles) | 10 |
| CTATAGCCATTATCTTTGCA | ||||
| iceA2 | GTTGGGTATATCACAATTTAT | 229 | 95°C 1 min; 57°C, 1 s; 72°C, 1 min (35 cycles) | 10 |
| TTTCCCTATTTTCTAGTAGGT | ||||
| babA2 | CCAAACGAAACAAAAAGCGT | 271 | 94°C, 1 min; 45°C, 1 min; 72°C, 1 min (30 cycles) | 10 |
| GCTTGTGTAAAAGCCGTCGT | ||||
| oipA | GTTTTTGATGCATGGGATTT | 401 | 94°C, 1 min; 56°C, 1 min; 72°C, 1 min (35 cycles) | 5 |
| GTGCATCTCTTATGGCTTT |
Statistical analysis
The Chi square and Fischer’s exact tests were applied to estimate the statistical differences between disease and various genotypes. The P-value < 0.05 considered as significant statistical differences.
RESULTS
The studied patients were 99 (44.6%) males and 123(55.4%) females with age range of 16 to 88 years old (mean age 44.6 ± 18.01 years). Clinical data revealed that from 222 patients with gastric complaints investigated by gastric endoscopy, 129 (58.1%) had gastritis, 47(21.2%) diagnosed as non-ulcer dyspepsia (NUD), 31 (13.9%) and 12 (5.4%) had gastric and duodenal ulcers, respectively. Three (1.4%) of patients had gastric carcinoma. The isolation rate of H. pylori strains from gastric biopsies was 37 out of 222 (16.7%), (95% CI: 0.118-0.216) in 12 males (32.4%), and 25 females patients (67.6%). The most common clinical diagnoses were non-ulcer dyspepsia in 29 patients (78.4%), followed by peptic ulcer diseases in 8 patients (21.6%). H. pylori DNA were extracted from 37 strains. DNA reliability and specificity was confirmed by glmM amplification and all of our isolates were positive for the glmM. The predominant genotype in strains by PCR was the babA2 (94.6%) followed by the oipA (81.1%), cagA (62.2%), the iceA1 (48.6%), and cagE in 40.5%, whereas the iceA2 was detected only in 16.2% and of strains.
The vacA alleles s1, s2, m1 and m2 were detected in 20 (54.1%), 13 (35.1%), 9 (24.3%) and 23 (62.2%) isolates, respectively. The allele s1m2 was the most frequent vacA allelic combination in H. pylori strains examined, followed by s2 m2, s1m1 and s2m1. The vacA s1a subtype was identified in H.pylori strains from 54.1% of patients (20 out of 37). The vacA s1b subtype was not recognized in this study, while the VacA s1c subtype was identified in 70.3% patients (26 out of 37), and the VacA s2 type was identified in 13 patients (35.1%). Mixed H. pylori VacA s1a and vacA s1c subtypes were present in 35.1%, and combination of vacA s2 and vacA s1c subtypes in 29.7% of patients (Table 2). There was no statistical association between vacAs1m1 and vacAs1m2 with clinical presentation. The iceA1 and iceA2 subtypes were detected in 48.6% and 16.2% of H. pylori infected patients, respectively. The iceA1 was found in 2 out of 8 (25%) of the peptic ulcer patients, while IceA2 was found in 55.2% (16 out of 29) of the non-ulcer dyspeptic patients. The babA2 gene was detected in 94.6% (35 out of 37) of the H. pylori infected patients. However, there was no statistically significant difference in each individual genes among the patient groups (p>0.05). The association of vacA with cagA, cagE, iceA1, iceA2, babA2, and oipA genotypes in 37 strains of H. pylori is presented in Table 2. The strains typed as vacAs1/cagA/baba2 detected in 43.2% and vacAs1/cagA/oipA in 40.5% of isolates (Fig. 1). A significant correlation was observed between vacAs1 and cagA genotypes (P<0.008), vacAs1/cagE (P = 0.001), vacAs2/cagA (P<0.047), and vacAs2/cagE (P = 0.016) with non-ulcer dyspepsia; but there were not observed any correlation between other virulence markers. The majority (96.6%) of patients with non-ulcer dyspepsia possessing the babA2 genotype and 86.2% defined as oipA genotype (Table 3). Distribution of H. pylori genotypes and risk for upper gastrointestinal diseases is showed in Table 4.
Table 2.
The percent of vacA with cagA, cagE, iceA1, iceA2, babA2, and oipA genotypes in 37 strains of H. pylori.
| vacA | No. (%) | cagA+ (%) | CagE+ ( %) | iceA1+ (%) | iceA2+ (%) | BabA+ (%) | OipA+ (%) |
|---|---|---|---|---|---|---|---|
| s-region | |||||||
| S1 | 20 (54.1 ) | 18 (90) | 16 (80) | 12 (60) | 3 (15) | 18 (90) | 17 (85) |
| S1a | 20 (54.1) | 18 (90) | 13 (65) | 13 (65) | 3 (15) | 18 (90) | 16 (80) |
| S1c | 26 (70.3) | 15 (57.7) | 10 (38.5) | 15 (57.7) | 6 (23.1) | 25 (96.2) | 21 (80.8) |
| S2 | 13 (35.1 ) | 4 (30.8) | 1 (7.7) | 6 (46.2) | 3 (23.1) | 13 (100) | 10 (76.9) |
| S1a+s1c | 13 (35.1) | 12 (92.3) | 9 (69.2) | 10 (76.9) | 3 (23.1) | 12 (92.3) | 10 (76.9) |
| S1c + S2 | 11 (29.7) | 3 (27.3) | 0 (0) | 6 (54.5) | 3 (27.3) | 11 (100) | 9 (81.8) |
| m-region | |||||||
| m1 | 9 (24.3) | 7 (77.8) | 5 (55.6) | 6 (66.7) | 3 (33.3) | 9 (100) | 8 (88.9) |
| m2 | 23 (62.2 ) | 16 (69.6) | 9 (39.1) | 12 (52.2) | 3 (13) | 21 (91.3) | 19 (82.6) |
| m1m2 | 1 (2.7) | 1 (100) | 1 (100) | 1 (100) | 0 (0) | 1 (100) | 1 (100) |
| s\m region | |||||||
| s1m1 | 6 (16.2) | 6 (100) | 5 (83.3) | 6 (100) | 2 (33.3) | 6 (100) | 6 (100) |
| s1m2 | 15 (40.5 ) | 13 (86.7) | 9 (60) | 7 (46.7) | 1 (6.7) | 13 (86.7) | 12 (80) |
| s2m1 | 2 (5.4) | 1 (50) | 0 (0) | 0 (0) | 1 (50) | 2 (100) | 1 (50) |
| s2m2 | 8 (21.6 ) | 3 (37.5) | 0 (0) | 5 (62.5) | 2 (25) | 8 (100) | 7 (87.5) |
Fig. 1.
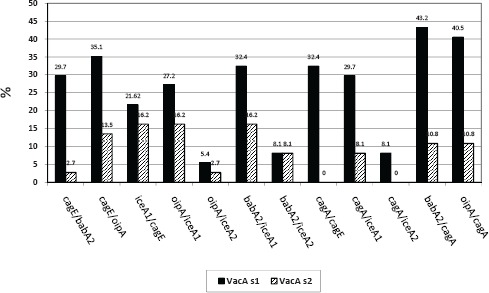
The percent of VacAs1 and VacAs2 in comparison with CagA+, CagE+, IceA1+, IceA2+, and BabA2+ genes of H. pylori; the bars represent the percent of persons infected with H. pylori having vacA genotype.
Table 3.
The clinical presentation of H. pylori infection according to H. Pylori strains genotype in studied patients.
| H. Pylori genotype Clinical presentation | Peptic ulcer (n = 8) NO. | non-ulcer dyspepsia (n = 29)NO. | Total NO. (%) |
|---|---|---|---|
| Vac A | |||
| m1 | 1 | 8 | 9 (24.3) |
| m2 | 6 | 17 | 23 (62.2) |
| m1m2 | 0 | 1 | 1 (2.7) |
| s1 | 4 | 16 | 20 (54.1) |
| s1a | 5 | 15 | 20 (54.1) |
| s1c | 6 | 20 | 26 (70.3) |
| s2 | 3 | 10 | 13 (35.1) |
| s1m1 | 0 | 6 | 6 (16.2) |
| s1m2 | 4 | 11 | 15 (40.5) |
| s2m1 | 1 | 1 | 2 (5.4) |
| s2m2 | 2 | 6 | 8 (21.6) |
| s1a +s1c | 3 | 10 | 13 (35.1) |
| s1c + s2 | 3 | 8 | 11 (29.7) |
| + cagA | 4 | 19 | 23 (62.2) |
| + cagE | 3 | 12 | 15 (40.5) |
| + iceA1 | 2 | 16 | 18 (48.6) |
| + iceA2 | 2 | 4 | 6 (16.2) |
| iceA1+ iceA2 | 0 | 3 | 3 (8.1) |
| + babA2 | 7 | 28 | 35 (94.6) |
| oipA | 5 | 25 | 30 (81.1) |
Table 4.
Distribution of H. pylori genotype and risk of upper gastrointestinal diseases.
| Genotypes | Peptic ulcer NO. (%) | Non ulcer dyspepsia NO. (%) | P value | Odds Ratio | Confidence Interval 95% Lower-Upper |
|---|---|---|---|---|---|
| m1 | 1 (11.1) | 8 (88.9) | 0.357 | 0.375 | 0.40-3.551 |
| m2 | 6 (26.1) | 17 (73.9) | 0.340 | 2.118 | 0.363-12.342 |
| s1 | 4 (20) | 16 (80) | 0.553 | 0.813 | 0.169-3.895 |
| s1a | 5 (25) | 15 (75) | 0.447 | 1.556 | 0.312-7.551 |
| s1c | 6 (23.1) | 20 (76.9) | 0.556 | 1.350 | 0.227-8.031 |
| s2 | 3 (21.4) | 11 (78.6) | 0.635 | 1.036 | 0.207-5.198 |
| cagA | 4 (17.4) | 19 (82.6) | 0.343 | 0.526 | 0.108-2.564 |
| cagE | 3 (20) | 12 (80) | 0.588 | 0.850 | 0.170-4.256 |
| babA2 | 7 (20) | 28 (80) | 0.390 | 0.250 | 0.014-4.511 |
| oipA | 5 (16.7) | 25 (83.3) | 0.156 | 0.267 | 0.045-1.579 |
| iceA1 | 2 (11.1) | 16 (88.9) | 0.133 | 0.271 | 0.047-1.576 |
| iceA2 | 2 (33.3) | 4 (66.7) | 0.387 | 2.083 | 0.306-14.168 |
DISCUSSION
It is known that more than half of the world’s human population is colonized by H. pylori (8). The predominant genotypes in this study were the babA2 followed by the oipA, cagA, iceA1, cagE and iceA2. Previous studies reported that the vacA genotype and occurrence of the cagA gene varied in H. pylori isolates collected from different parts of the world. These genotype variations affect the clinical presentation in H. pylori infected patients. The presence of the cagA gene varies from a minimum of 50% in some Middle East (9) to a maximum of 99% in many East Asian countries (10,11). The percentage of cagA-positive H. pylori strains found in our study is less than reported data from European and North American studies (74% to 88%) (12-14). Many studies have proposed that cagA is a useful marker for the most virulent strains that are associated with peptic ulcer diseases, atrophic gastritis and adenocarcinoma (15). The vacA is an important virulence factor in nearly half of H. pylori isolates that encoding the vacuolating cytotoxin in various mammalian cell lines in-vitro. The H. pylori isolates classified according to presence of different families of vacA signal sequences (s1a, s1b, s2), and middle region alleles (m1, m2) (16). In this study vacA s1 was seen more than vacAs2. The study from Middle East showed that vacA s1 and s2 genotypes were similarly expected to be present in patients. African Arabs mainly were infected with s2 and South-Asians with the s1 genotypes (9). In the present study the vacA s1m2 genotype was found in 40.5% and vacA s2m2 in 21.6% of H. Pylori infected patients. According to previous studies, the most frequent genotype vacA s1m2, isolated in this study had lower vacuole formation activity than the vacAs1m1, which might accomplished by less severe pathological effects as well as less clinical consequences (40.5% vs. 16.2%), while those with vacAs2 (35.1%) fails to induce cell vacuolation in-vitro. Strains carrying the s1ml mosaic combination of the gene vacA show higher levels of cytotoxic activity than s1m2 strains, whereas s2m2 strains do not secret the vacuolating cytotoxin. The m1vacA and m2vacA, which are mostly formed by isolates containing the s1/m1vacA and s1/m2vacA genes, respectively; have different cell type specificities in cytotoxicity study (16). The vacAs1c genotype was dominant in this study (70.3%), but the vacA s1b subtype was not recognized. The presence of multiple organisms within a host may occur as a result of recombination procedures leading to genetic shift, however ongoing mutation inside a strain may leads to the formation of quasi species by genetic drift. Several genotypic markers such as cagA, vacA, s1a and iceA1 are related with an increased risk of disease (17). The iceA gene may be related with peptic ulcer disease (18) while some studies have recommended a contrary association (19). The iceA1 genotype detected in 48.6% of our patients. This finding agrees with previous reports that the IceA1 allele was found more frequent than the IceA2 allele in Chinese, Japanese, Korean, Dutch and Thai Patients (10, 20-22). The iceA2 has been found to be main allele among American and Brazilian patients (19,22). The prevalence rate for BabA2 in this study was94.6%, which is higher than reports from Colombia 57% and Costa Rica 73.7%, but it is similar to results from Chilie 97.4% and Japan 96.8% (23-25). In a study from Isfahan, Iran, the incidence rate of babA2 was 71.6%. They reported there is no relationship between genotype and clinical outcomes (gastritis and PUD) (25). Most of the H. pylori strains in Asia are babA2 positive, surprisingly unrelated to clinical outcome (26). This study revealed a high prevalence of oipA genes (81.1%), which is in agreement with the prevalence of oipA genes strains in Bulgarian patients (27), but is far less than the data reported from Tunisia (90.8%) (5). In present survey the oipA gene was found in 62.5% of peptic ulcer patients and 86.2% of non-ulcer dyspepsia. Significant correlations were observed between vacAs1/vacAs2 with cagA and cagE genotypes. On the other hand, we did not observe any correlation between vacAs1 and iceA1, iceA2, oipA and babA2 genotypes. No significant relationship was observed between vacA genotypes and the manifestations of peptic ulcer diseases, which is in agreement with previous reports (28). Ribeiro et al. showed that vacAs1 was the only prognostic factor for peptic ulcer disease (29). Molaei et al. reported that the frequencies of vacA gene subtypes s1, s2, m1 and m2 in 78 isolated strains were 70.5%, 29.5%, 37.2% and 62.8%, respectively. They showed that vacAs1 was significantly associated with more severe gastritis and 83.3% of the vacA-positive strains had s1 allele (30). Dabiri et al. reported that there was no significant association between cagA and cagE status or vacA genotypes and clinical outcomes. The oipA-positive strains were more common in nonulcer dyspepsia than in peptic ulcer patients (6). The present study showed that patients with peptic ulcer disease and non-ulcer dyspeptic patients nearly were infected equally by multiple strains of H. pylori. The strains typed as vacA s1/ cagA+/ iceA+/ oipA+, and babA2 + were more prevalent than those typed as vacAs2/ cagA+/ iceA+/ oipA+, and babA2+. With close concern to distribution of virulence genes to sex, educational status, and smoking habit there were no statistically significant difference among vacA, cagA, cagE, iceA, babA2, and oipA genes. No significant correlation were found between the existence of vacA, cagA, cagE, iceA, babA2, and oipA genes with peptic ulcer diseases groups in studied patients.
In conclusion, this is the first study that reveals a high prevalence of baba2, oipA, vacA, cagA, iceA1 and cagE genes in H. pylori isolates in Kashan. The s1m2 genotype was the most prevalent among all patients, but there was no correlation to peptic ulcer disease. Statistically significant correlations were observed between vacAs1/cagA, vacAs1/cagE, vacAs2/cagA and vacAs2/cagE virulence markers with Non-ulcer dyspepsia..
Acknowledgments
This project was supported by Kashan University of Medical Sciences Research fund. We are grateful to Dr. Kamran Dastehgoli for editing the article.
References
- 1.Suzuki R, Shiota S, Yamaoka Y, et al. Molecular epidemiology,population genetics, and pathogenic role of Helicobacter pylori. Infect Genet Evol. 2012;12:203–213. doi: 10.1016/j.meegid.2011.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tanih NF, McMillan M, Naidoo N, Ndip LM, Weaver LT, Ndip RN, et al. Prevalence of Helicobacter pylori vacA,cagA and iceA genotypes in South African patients with upper gastrointestinal diseases. Acta Trop. 2010;116:68–73. doi: 10.1016/j.actatropica.2010.05.011. [DOI] [PubMed] [Google Scholar]
- 3.Yamaoka Y, Kikuchi S, el-Zimaity HM, Gutierrez O, Osato MS, Graham DY, et al. Importance of Helicobacter pylori oipA in clinical presentation, gastric inflammation, and mucosal interleukin 8 production. Gastroenterology. 2002;123:414–424. doi: 10.1053/gast.2002.34781. [DOI] [PubMed] [Google Scholar]
- 4.Paniagua GL, Monroy E, Rodríguez R, Arroniz S, Rodríguez C, Cortés JL. Frequency of vacA, cagA and babA2 virulence markers in Helicobacter pylori strains isolated from Mexican patients with chronic gastritis. Ann Clin Microbiol Antimicrob. 2009;30:8–14. doi: 10.1186/1476-0711-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ben Mansour K, Fendri C, Zribi M, Masmoudi A, Labbene M, Fillali A, et al. Prevalence of Helicobacter pylori vacA, cagA, iceA and oipA genotypes in Tunisian patients. Ann Clin Microbiol Antimicrob. 2010;19:9–10. doi: 10.1186/1476-0711-9-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dabiri H, Maleknejad P, Yamaoka Y, Feizabadi MM, Jafari F, Rezadehbashi M, et al. Distribution of Helicobacter pylori cagA, cagE, oipA and vacA in different major ethnic groups in Tehran, Iran. J Gastroenterol Hepatol. 2009;24:1380–1386. doi: 10.1111/j.1440-1746.2009.05876.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mohammadi M, Oghalaie A, Mohajerani N, Massarrat S, Nasiri M, Bennedsen M. Prevalence of Helicobacter pylori vacuolating cytotoxin and its allelic mosaicism as a predictive marker for Iranian dyspeptic patients. Bull Soc Pathol Exot. 2003;96:3–5. [PubMed] [Google Scholar]
- 8.Atherton JC. The pathogenesis of Helicobacter pyloriinduced gastro-duodenal diseases. Annu Rev Pathol. 2006;1:63–96. doi: 10.1146/annurev.pathol.1.110304.100125. [DOI] [PubMed] [Google Scholar]
- 9.Al Qabandi A, Mustafa AS, Siddique I, Khajah AK, Madda JP, Junaid TA, et al. Distribution of vacA and cagA genotypes of Helicobacter pylori in Kuwait. Acta Trop. 2005;93:283–288. doi: 10.1016/j.actatropica.2005.01.004. [DOI] [PubMed] [Google Scholar]
- 10.Chomvarin C, Namwat W, Chaicumpar K, Mairiang P, Sangchan A, Sripa B. Prevalence of Helicobacter pylori vacA, cagA, cagE, iceA and babA2 genotypes in Thai dyspeptic patients. Int J Infect Dis. 2008;12:30–36. doi: 10.1016/j.ijid.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 11.Lai CH, Kuo CH, Chen YC, Chao FY, Poon SK, Chang CS, et al. High prevalence of cagA- and babA2-positive Helicobacter pylori clinical isolates in Taiwan. J Clin Microbiol. 2002;40:3860–3862. doi: 10.1128/JCM.40.10.3860-3862.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Miehlke S, Kibler K, Kim JG, Figura N, Small SM, Graham DY. Allelic variation in the cagA gene of Helicobacter pylori obtained from Korea compared to the United States. Am J Gastroenterol. 1996;91:1322–1325. [PubMed] [Google Scholar]
- 13.Van Doorn LJ, Figueiredo C, Mégraud F, Pena S, Midolo P, Queiroz DM, et al. Geographic distribution of vacA allelic types of Helicobacter pylori. Gastroenterology. 1999;116:823–830. doi: 10.1016/s0016-5085(99)70065-x. [DOI] [PubMed] [Google Scholar]
- 14.Yamaoka Y, Kodama T, Gutierrez O, Kim JG, Kashima K, Graham DY. Relationship between Helicobacter pylori iceA, cagA, and vacA status and clinical outcome:studies in four different countries. J Clin Microbiol. 1999;37:2274–2279. doi: 10.1128/jcm.37.7.2274-2279.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Watada M, Shiota S, Matsunari O, Suzuki R, Murakami K, Fujioka T, et al. Association between Helicobacter pylori cagA-related genes and clinical outcomes in Colombia and Japan. BMC Gastroenterol. 2011 Dec 22;:11–141. doi: 10.1186/1471-230X-11-141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Isomoto H, Moss J, Martin D, Hirayama T. Pleiotropic actions of Helicobacter pylori vacuolating cytotoxin, VacA. Tohoku J Exp Med. 2010 Jan;220:3–14. doi: 10.1620/tjem.220.3. [DOI] [PubMed] [Google Scholar]
- 17.Blaser MJ. Heterogeneity of Helicobacter pylori. Eur J Gastroenterol Hepatol. 2012;9(Suppl 1):S3–6. doi: 10.1097/00042737-201204001-00002. discussion S6-7. [DOI] [PubMed] [Google Scholar]
- 18.Podzorski RP, Podzorski DS, Wuerth A, Tolia V. Analysis of the vacA, cagA, cagE, iceA, and babA2 genes in Helicobacter pylori from sixty-one pediatric patients from the Midwestern United States. Diagn Microbiol Infect Dis. 2003;46:83–88. doi: 10.1016/s0732-8893(03)00034-8. [DOI] [PubMed] [Google Scholar]
- 19.Kim SY, Woo CW, Lee YM, Son BR, Kim JM, Chae HB. Genotyping CagA, VacA subtype, IceA1, and BabA of Helicobacter pylori isolates from Korean patients, and their association with gastroduodenal diseases. J Korean Med Sci. 2001;16:579–584. doi: 10.3346/jkms.2001.16.5.579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Han YH, Liu WZ, Zhu HY, Xiao SD, et al. Clinical relevance of iceA and babA2 genotypes of Helicobacter pylori in a Shanghai population. Chin J Dig Dis. 2004;5:181–185. doi: 10.1111/j.1443-9573.2004.00175.x. [DOI] [PubMed] [Google Scholar]
- 21.Ashour AA, Collares GB, Mendes EN, de Gusmão VR, Queiroz DM, Magalhães PP. iceA genotypes of Helicobacter pylori strains isolated from Brazilian children and adults. J Clin Microbiol. 2001;39:1746–1750. doi: 10.1128/JCM.39.5.1746-1750.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Arévalo-Galvis A, Trespalacios-Rangell AA, Otero W, Mercado-Reyes MM, Poutou-Piñales RA. Prevalence of cagA, vacA, babA2 and iceA genes in H. pylori strains isolated from Colombian patients with functional dyspepsia. Pol J Microbiol. 2012;61:33–40. [PubMed] [Google Scholar]
- 23.González I, Romero J, Rodríguez B, Llanos J, Morales E, Figueroa H, et al. High prevalence of virulenceassociated genotypes in Helicobacter pylori clinical isolates in the Region del Maule, Chile. Scand J Infect Dis. 2011;43:652–655. doi: 10.3109/00365548.2011.572909. [DOI] [PubMed] [Google Scholar]
- 24.Con SA, Takeuchi H, Nishioka M, Morimoto S, Sugiura T, Yasuda N, et al. Clinical relevance of Helicobacter pylori babA2 and babA2/B in Costa Rica and Japan. World J Gastroenterol. 2010;28(16):474–478. doi: 10.3748/wjg.v16.i4.474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ghasemian Safaei H, Havaei SA, Tavakkoli H, Eshaghei M, Navabakbar F, Salehei R. Relation of babA2 genotype of Helicobacter pylori infection with chronic active gastritis, duodenal ulcer and non-cardia gastric cancer in Alzahra hospital, Isfahan, Iran. JJM. 2010;3:93–98. [Google Scholar]
- 26.Mizushima T, Sugiyama T, Komatsu Y, Ishizuka J, Kato M, Asaka M, et al. Clinical relevance of the babA2 genotype of Helicobacter pylori in Japanese clinical isolates. J Clin Microbiol. 2001;39:2463–2465. doi: 10.1128/JCM.39.7.2463-2465.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Markovska R, Boyanova L, Yordanov D, Gergova G, Mitov I. Helicobacter pylori oipA genetic diversity and its association with both diseases and cagA, vacA s, m, and i alleles among Bulgarian patients. Diagn Microbiol Infect Dis. 2011;71:335–340. doi: 10.1016/j.diagmicrobio.2011.08.008. [DOI] [PubMed] [Google Scholar]
- 28.Faundez G, Troncoso M, Figueroa G. cagA and vacA in strains of Helicobacter pylori from ulcer and nonulcerative dyspepsia patients. BMC Gastroenterol. 2002 Sep 10;:2–20. doi: 10.1186/1471-230X-2-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ribeiro ML, Godoy AP, Benvengo YH, Mendonça S, Pedrazzoli J, Jr., et al. Clinical relevance of the cagA, vacA and iceA genotypes of Helicobacter pylori in Brazilian clinical isolates. FEMS Immunol Med Microbiol. 2003;36(25):181–185. doi: 10.1016/S0928-8244(03)00029-4. [DOI] [PubMed] [Google Scholar]
- 30.Molaei M, Foroughi F, Mashayekhi R, Haghazali M, Zojaji H, Jafari F, Dabiri H, Zali MR. CagA status and VacA subtypes of Helicobacter pylori in relation to histopathologic findings in Iranian population. Indian J Pathol Microbiol. 2010 Jan-Mar;53:24–27. doi: 10.4103/0377-4929.59178. [DOI] [PubMed] [Google Scholar]


