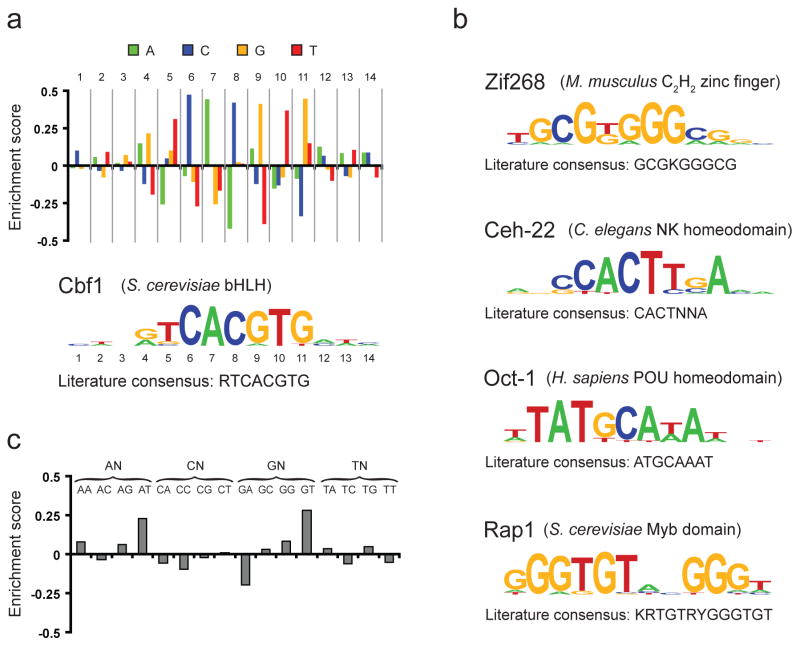
Figure 3.
Determination of motifs and logos for five TFs. (a) Method of constructing PWMs and sequence logos, using Cbf1 as an example. First, all 8-mers containing up to three gapped positions are evaluated using our enrichment score (see Methods), and the highest-scoring 8-mer (in this case GTCACGTG) is used as a seed for constructing the motif. Second, at each position within this 8-mer seed, all four possible nucleotides are compared by inspecting the ranks of the probes matching each of the four variants. This analysis produces a score between −0.5 and 0.5 for each variant at each position. Third, positions outside the 8-mer seed are inspected by dropping the least informative position within the seed and repeating the preceding analysis at every additional position that yields an 8-mer with at most three gaps (ensuring that the positions inspected outside of the 8-mer seed are based on a roughly equal number of samples to those within the 8-mer seed). This analysis produces the bar graph shown. Finally, these values are converted into a sequence logo by utilizing a suitably scaled Boltzmann distribution (see Supplementary Methods). (b) Logos for four additional TFs constructed using this method. For each, the organism and structural class are given. Consensus sequences in panels (a) and (b) were obtained from the literature for Cbf1 (ref. 27), Zif268 (ref. 28), Ceh-22 (ref. 29), Oct-1 (ref. 30), and Rap1 (ref. 12) (standard IUPAC abbreviations are used (K={T,G}; R={A,G}; Y={C,T}; N={A,C,G,T}). (c) Extension of the method for motif construction described in panel (a) to the case of di-nucleotide variants and applied to the first two positions in the Cbf1 motif. Here, all 16 variants of the form NNCACGTG were obtained and the enrichment score of each was computed.