Abstract
Extraction of medication information embedded in clinical text is important for research using electronic health records (EHRs). However, most of current medication information extraction systems identify drug and signature entities without mapping them to standard representation. In this study, we introduced the open source Java implementation of MedEx, an existing high-performance medication information extraction system, based on the Unstructured Information Management Architecture (UIMA) framework. In addition, we developed new encoding modules in the MedEx-UIMA system, which mapped an extracted drug name/dose/form to both generalized and specific RxNorm concepts and translated drug frequency information to ISO standard. We processed 826 documents by both systems and verified that MedEx-UIMA and MedEx (the Python version) performed similarly by comparing both results. Using two manually annotated test sets that contained 300 drug entries from medication list and 300 drug entries from narrative reports, the MedEx-UIMA system achieved F-measures of 98.5% and 97.5% respectively for encoding drug names to corresponding RxNorm generic drug ingredients, and F-measures of 85.4% and 88.1% respectively for mapping drug names/dose/form to the most specific RxNorm concepts. It also achieved an F-measure of 90.4% for normalizing frequency information to ISO standard. The open source MedEx-UIMA system is freely available online at http://code.google.com/p/medex-uima/.
INTRODUCTION
Electronic Health Records (EHRs) are becoming an enabling resource for drug outcome studies.1 However, medication data are often recorded in heterogeneous formats in EHRs. With the increased use of computerized provider order entry (CPOE) systems, electronic prescribing (e-prescribing) tools, and electronic medication administration record systems (e-MARs), medication records in the EHR are increasingly available as structured entries. However, much current and historical medication information is still embedded in narrative text entries within clinical documentation, patient problem lists, or communications with patients through telephone calls or patient portals, especially in the outpatient settings. Therefore, natural language processing (NLP) methods that can extract medication information from clinical narratives and encode them into standard representations have received great attention, as detailed below.
Early studies primarily focused on extracting drug names from clinical notes. In 1996, Evans et al. built the CLARIT2 system to extract the drug name and dosage phrases in discharge summaries and reported an accuracy of 80%. Chhieng et al.3 reported a precision of 83% by using a string matching method to identify drug names in clinical records. In 2009, Jagannathan et al.4 evaluated the performance of four commercial clinical NLP systems on medication information extraction (including drug names, strength, route, and frequency). These systems demonstrated high F-measures (93.2%) for capturing drug names, but lower F-measures (85.3%, 80.3%, and 48.3% respectively) on retrieving strength, route, and frequency. In 2009, Informatics for Integrating Biology and the Bedside (i2b2), an NIH-funded National Center for Biomedical Computing (NCBC) based at Partners Healthcare System in Boston, organized an clinical NLP challenge to extract medication names and their associated signature fields including dosage, mode, frequency, duration, and reason from hospital discharge summaries.5 Twenty teams from twenty-three organizations and nine countries participated in the challenge. A variety of medication information extraction systems were developed and included systems using rule-based,6 machine learning based,7,8 and hybrid approaches,9 with overall promising results.
Despite the active NLP work on medication extraction, most of existing systems output medication related entities as textual fields, without mapping to standard representations such as RxNorm10 for drugs and ISO 8601 standard for frequency information. One study done by Levin and colleagues11 developed an effective rule-based system to extract drug names from anesthesia records and map to RxNorm concept unique identifiers (RxCUIs), with 92.2% sensitivity and 95.7% specificity. However, this study focused on encoding drug ingredients/brands only. In the example “Cetirizine 5 mg oral tablet”, Levin’s system will only encode the drug name “Cetirizine” (RxCUI 20610). However, an RxNorm concept actually can include three components: drug name (generic or brand), dose, and form. For the above example, a more specific RxCUI (1014676 – “cetirizine hydrochloride 5 MG Oral Tablet”) could be assigned. With available drug dose and form (and/or route) information extracted by NLP systems, more specific RxCUIs can be assigned to medications in clinical text, which can be useful for other computerized applications. For example, the dose form (e.g., intravenous vs. oral vs. topical) can imply very different indications and side effects. Frequency information is also important for medications and different string variants can often represent the same frequency (e.g., “two times a day” is equivalent to “b.i.d”). Therefore, normalization of drug frequency information is needed. However, few clinical NLP systems provide normalized frequency values. In the 2012 i2b2 NLP challenge on temporal information extraction, temporal expressions including frequency were normalized based on the ISO 8601 standard as in the TIMEX312 tag, which is the part of TimeML, a formal specification language for events and temporal expressions. To the best of our knowledge, TIMEX3 normalization has not been applied to the extraction of drug frequency information in clinical text.
In previous work, we developed MedEx,13 a Python-based NLP system which could extract drug names and signature information with over 90% F-measure in discharge summaries and clinical visit notes from Vanderbilt University Hospital. We applied an extended version of MedEx to the 2009 i2b2 NLP challenge on medication extraction; it was ranked as the second best system among twenty entries.6 We also developed simple normalization modules for dose and frequency, and integrated them with MedEx to calculate daily dose of tacrolimus14 and weekly dose of warfarin.15 In this study, we re-implemented MedEx in Java, based on the Unstructured Information Management Architecture (UIMA),16 which is a component software architecture for development, discovery, composition, and deployment of multi-modal analytics for unstructured data. We name the new system “MedEx-UIMA” and it is freely available as open-source software. We also developed two new components in MedEx-UIMA and evaluated them herein: 1) encoding drugs to specific RxNorm concepts and 2) normalizing frequency to TIMEX3 format.
METHODS
As shown in Figure 1, the MedEx-UIMA system consists of two main components: 1) an information extraction module, which extracts medication related fields from clinical text; and 2) a standardization module that encodes drug name/dose/form information into RxCUIs and normalizes frequency information to the TIMEX3 format. The information extraction module basically is a Java implementation of the previous Python version of MedEx, with additional changes in transformation and disambiguation. The RxCUI encoding and frequency normalization are new functionalities of MedEx-UIMA. They are the primary focus of this study.
Figure 1.
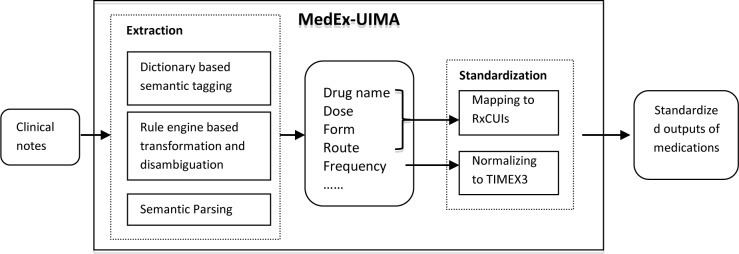
An overview of the MedEx-UIMA system
The UIMA implementation of MedEx
Using on the UIMA framework, we re-built the MedEx in Java as a pipeline-based system, where we defined classes including Sentence Boundary Detector, Tokenizer, Section Tagger, Semantic Tagger, Parser and Encoder. One significant change to the new MedEx-UIMA system is that we applied the Drools rule engine (http://www.jboss.org/drools/) to handle heuristic rules used in semantic tagging for tag transformation and word sense disambiguation. The rule-engine separates rule management from the workflow, thus making it possible for non-technical users to modify rules needed for specific tasks. The encoder is a new component in MedEx-UIMA, which maps drug name, dose, and form information to most specific RxNorm concepts and normalizes frequency information to TIMEX3 format.
Mapping drug name, dose, and form information to RxNorm concepts
When encoding drug information extracted from clinical text using RxNorm, there are two primarily options: 1) least-specific: map drug names only, e.g., to generic names such as “cetirizine”; or 2) most-specific: map to more specific RxNorm concepts that could contain drug name (either generic or brand), dose, and form information, such as “Cetirizine 5 MG Oral Tablet.” In MedEx-UIMA, we provide both types of RxCUIs for a given drug entity. It is straightforward to map drug names to least specific RxCUIs (the generic ingredient). We created a mapping between brand names and generic names based on RxNorm relationships and built a simple dictionary lookup function to map extracted drug names to their corresponding generic name RxCUIs.
Determining the most specific RxCUIs based on extracted drug name, dose, route, and form information is more challenging. We developed a rule-based approach for this task, which consists of four steps:
Normalize drug information extracted by MedEx: Five fields extracted by MedEx including drug name, dose, dose amount, route, and form are used to generate normalized fields of drug name, dose, and form. The normalization process is based on heuristic rules and manually created knowledge bases. In the example of “Cetirizine 5000 mcg tabs”, MedEx will recognize “cetirizine” as a drug name, “5000 mcg” as the dose, and “tablet” as the form. The normalization program will produce normalized results as (Generic name: cetirizine), (Dose: 5 mg), and (Form: tablet), which can then be mapped to the RxNorm entry. In this example, rules for conversion between different units in the dose field and knowledge for recognizing “Tab” and “Tablet” as synonyms were used in the normalization process. We have developed knowledge bases about synonyms and route-form mappings for normalizing drug forms.
Normalize drug information of RxNorm concepts: For each RxNorm term, we process it using the same procedure as in step 1 and generate normalized fields for drug names, dose, form etc.
Generate RxNorm candidate entries: For a given drug entry, we search all RxNorm concepts and generate a list of candidate concepts containing the same normalized drug name.
Rank RxNorm candidate concepts by calculating similarity scores between the normalized drug entry and candidate concepts: Once the drug name, dose and form information is normalized, we concatenate them in an order to generate a string. We then calculate weighted Jaccard Similarity17 scores between a drug entry string and all its corresponding candidate string. The Jaccard Similarity is defined as the ratio between the number of common words in both two strings, multiplied with the weight of each word, and the number of words in any of two strings, multiplied with their weight. We assign different weights to different drug fields to reflect their search priorities. For example, the default weight of any word is “1”. But we assign a higher weight (e.g., 1.8) to the dose field, as the same dose is a strong indicator. The RxNORM concept with the highest similarity score with the drug entry is then selected as the most specific RxNORM code.
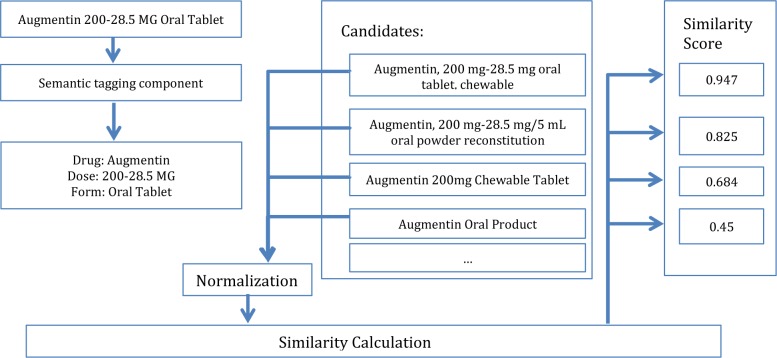
Figure 2 shows an example of searching the most specific RxNORM codes. As shown in the figure, “Augmentin 200-28.5 MG Oral Tablet” is the input sentence. Drug (Augmentin), dose (200-28.5 MG) and form (Oral Table) are extracted and normalized by MedEx. After searching the drug name “Augmentin”, multiple RxNORM candidate entries are generated, including “Augmentin, 200 mg-28.5 mg oral tablet, chewable”, “Augmentin, 200 mg-28.5 mg/5 mL oral powder” etc. All RxNORM candidate entries are normalized in the same way. Then we calculate Jaccard similarity between the string “Augmentin 200-28.5 MG Oral Tablet” and each of the RxNORM candidate strings. The one with highest similarity score is then selected as the most specific RxNORM entry.
Figure 2.
The example of determination of most specific RxNORM code
Normalizing drug frequency information to the TIMEX3 format
The frequency normalization module was constructed on our temporal expression extraction system developed for the 2012 i2b2 NLP challenge on temporal information extraction. The original system is a rule-based system developed in Python to extract three types of temporal expressions, including date, frequency and duration. We re-implemented the system in Java and extended it with new regular expression rules for handling additional drug frequency patterns observed in the development set. The following example shows how the rule-based system normalizes frequencies into TIMEX3 format. For the expression “three times per week”, the frequency normalization module first detects the normalizable components using the rule “(%NumWord) (%TIMES) (%PER) (%DayUnit)”. Strings starting with “%” are predefined patterns using regular expressions, where “NumWord” is a lexicon of all the possible numbers in English words, “TIMES” is a lexicon of all the possible expression for times (e.g., “times”, “x”), “PER” is a lexicon of all the possible expression for every (e.g., “every”, “per”, “each”), and “DayUnit” is a lexicon of all the possible units of days (e.g., “day”, “week”, “month”). Once the regular expression is triggered, the normalization rules will be applied to normalize the NumWord “three” into “3”, DayUnit “week” into “W” to generate the normalized value “R3P1W”, where R stands for “Repeat” and P stands for “Period.” One difference between our drug frequency normalization and the i2b2 challenge guidelines was that we do not average a range (e.g., the i2b2 guidelines normalize “three to four weeks” into “P3.5W”; however, our system normalizes it as “P3-4W”)
Evaluation
We first compared the performance of the MedEx-UIMA with the previous Python-based MedEx system (MedEx-Python). We processed 826 clinical notes from the 2010 i2b2 challenge using both MedEx-Python and the MedEx-UIMA systems. We then took the outputs of MedEx-Python as the gold standard and calculated precision/recall/F-measure of MedEx-UIMA against the gold standard. In addition, we reviewed 100 randomly selected discrepant drug entities by the two systems and counted the number of correct samples by MedEx-UIMA.
To develop and evaluate the encoding modules for drug name and frequency information, we created manually annotated datasets. We first used the dataset from the 2009 i2b2 clinical NLP challenge, which was to extract medication information from discharge summaries. The i2b2 dataset contains 251 discharge summaries collectively annotated by challenge participants, in which drug names and associated strength, route and frequency information were identified. We randomly divided the dataset into two subsets: 126 notes as the development set and 125 notes as the test set. From the development set, we collected all i2b2 annotated drug entities and annotated 300 randomly-selected distinct drug entities. These 300 drug entities (with their sentences) were used to develop our system.
From the test set, we also collected all drug entities and randomly selected 300 drugs for annotation, which served as the independent test set to evaluate our system. For each drug entity in the development and test set, the original sentence containing the drug as well as drug name, dose, and route fields extracted by the i2b2 challenge, were presented to a medical domain expert for manual review. To encode RxNorm concepts, the annotator searched RxCUIs using RxNav, which is graphical search interface for RxNorm concepts. For frequency normalization, the annotator manually entered the normalized value for each frequency expression. In addition to the i2b2 dataset, which primarily contains drug entries in clinical narratives, we generated another test set containing more structured medication data. We randomly selected a list of 300 medications entries from computerized order entry system at UT Physician, a clinic of University of Texas Health Science Center at Houston, and manually annotated them with RxNORM codes following the same procedure.
We evaluated the performance of our system by reporting standard precision, recall, and F-measure on the independent test sets. For the first dataset, as the i2b2 challenge included drug classes such as “antibiotics”, not all 300 drug entities in the test set can be coded by RxNorm concepts. Based on manual review, 270 drugs in the test set were classified as codable drugs. Among 270 codable drugs, true positives were defined as samples that were extracted by MedEx-UIMA and assigned correct RxCUIs. Recall was defined as the ratio between the number of true positives and the total number of codable drugs (270). Precision was defined as the ratio between the number of true positives and the number of codable drugs recognized by MedEx-UIMA. Similar definitions were used to measure precision and recall for frequency normalization as well. There were 243 frequency expressions in the independent test set. To be qualified as true positives, a frequency expression must be recognized by MedEx-UIMA and assigned the correct normalized values in the TIMEX3 format. For the medication list from UT Physician, all three hundred medication entries were codable.
RESULTS
When the outputs of MedEx-Python served as gold standard, the MedEx-UIMA achieved a precision of 95.8%, a recall of 98.0%, and an F-measure of 96.9%, for recognizing all drug related fields including name, dose, route, frequency etc. Manual review of 100 discrepant results by two systems showed that 42% were judged better in MedEx (Python), and 58% were judged better in MedEx-UIMA. Thus, overall we estimate that MedEx-UIMA slightly outperforms the original version of MedEx in precision.
Table 1 shows the performance of MedEx-UIMA on extracting and encoding medication information using the independent test set. For mapping drug names to generic ingredients (least specific RxCUIs), on both the medication list and clinical narratives, the system achieved F-measure (98.5% and 97.5% respectively), which was consistent with previously reported high performance of MedEx on recognizing drug names. Mapping to the most-specific RxCUIs (taking dose and form into consideration) was more challenging: MedEx-UIMA achieved a precision of 85.8% and recall of 85.0% on drugs from medication list and 89.3% and 87.0% on drugs from clinical narratives. For frequency normalization, our system reached a high F-measure of 90.4% (precision 91.9% and recall 88.9%) on clinical narratives.
Table 1.
Evaluation results of MedEx-UIMA on extracting and encoding drug and frequency information
| Tasks | Precision | Recall | F-measure |
|---|---|---|---|
| Drug encoding - least-specific RxCUIs (Clinical narratives) | 98.8 % | 96.3 % | 97.5 % |
| Drug encoding - most specific RxCUIs (Clinical narratives) | 89.3% | 87.0% | 88.1 % |
| Frequency normalization (Clinical narratives) | 91.9% | 88.9% | 90.4% |
| Drug encoding - least-specific RxCUIs (Medication list) | 99.0% | 98.0% | 98.5% |
| Drug encoding - most-specific RxCUIs (Medication list) | 85.8% | 85.0% | 85.4% |
DISCUSSION
In this study, we re-implemented MedEx, a high performance medication information extraction system, in Java using the UIMA framework. Evaluation showed the MedEx-UIMA system had similar high performance on recognizing drug related entities as MedEx (Python version). We also extended the encoding function of MedEx-UIMA to map drug names to generic ingredients and also the most specific RxNorm concepts, and developed a module to normalize frequency expressions to the standard TIMEX3 format. Our evaluation using a test set from the 2009 i2b2 challenge demonstrated that MedEx-UIMA can extract and encode drug name and frequency information with good performance. Such standard medication information extracted from clinical text can not only facilitate EHR-based clinical and translational research, but can also benefit computerized clinical applications such as clinical decision support systems and medication reconciliation processes. More importantly, MedEx-UIMA is available to the public as an open-source system, which can be freely downloaded from Google Code at http://code.google.com/p/medex-uima/.
We analyzed errors in mapping drug name/dose/form to RxNorm Concepts. Recall errors were often caused by unrecognized synonyms, abbreviations, or misspelled words. For example, “MVI” is a common abbreviation for “Multi-Vitamins”; but it could not be mapped to the expanded name by MedEx-UIMA, thus no RxCUI could be assigned. Precision errors had two primary causes. One is related to insufficient rules or knowledge for normalizing drug name, dose, and form information extracted by the NLP system. For example, “regular insulin” was not mapped because we did not add the fact of “regular insulin” = “insulin” to our knowledge base. The other regards selecting the correct RxCUI from multiple candidate concepts. Our current approach relies on simple string matching between drug name, dose, and form fields. More sophisticated code selection methods will be investigated in future development. For example, we plan to look into information retrieval methods to rank candidate concepts based on the querying drug string.
This study has limitations. One of them is the annotation process, which only involved one annotator, with some oversight and review of unclear cases by a board-certified internist. We plan to recruit multiple annotators for future development so that we can reduce bias introduced by annotation. The evaluation of drug name encoding and frequency normalization was based on selected drug entities at sentence level. In the future, we plan to further evaluate the performance of MedEx-UIMA at the clinical document level. Another limitation is that only documents from the i2b2 challenge were used; future studies should examine more documents types from other institutions.
CONCLUSION
In this study, we developed MedEx-UIMA, an open source medication information extracting and encoding system based on the existing MedEx system. It not only recognizes medication related entities with high performance, but also encodes drug names to specific RxNorm concepts and frequency information to ISO standard. Such a tool will have broad uses in various clinical settings, as well as EHR-based clinical and translational research.
Acknowledgments
This study was supported in part by National Institute of General Medical Sciences grant 1R01GM102282, National Cancer Institute grant R01CA141307, Cancer Prevention & Research Institute of Texas grant RX1307, and the Office of the National Coordinator for Health Information Technology grant No. 10510592 for Patient-Centered Cognitive Support under the Strategic Health IT Advanced Research Projects Program (SHARP). We would like to thank organizers of the i2b2 clinical NLP challenges for providing the annotated data sets for research uses.
References
- 1.Wilke RA, Xu H, Denny JC, et al. The emerging role of electronic medical records in pharmacogenomics. Clinical pharmacology and therapeutics. 2011 Mar;89(3):379–386. doi: 10.1038/clpt.2010.260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Evans DA, Brownlow ND, Hersh WR, Campbell EM. Automating concept identification in the electronic medical record: an experiment in extracting dosage information. Proc AMIA Annu Fall Symp. 1996:388–392. [PMC free article] [PubMed] [Google Scholar]
- 3.Chhieng D, Day T, G G. Use of natural language programming to extract medication from unstructured electronic medical records. AMIA Annu Symp Proc. 2007;908 [PubMed] [Google Scholar]
- 4.Jagannathan V, Mullett CJ, Arbogast JG, et al. Assessment of commercial NLP engines for medication information extraction from dictated clinical notes. Int J Med Inform. 2009 Apr;78(4):284–291. doi: 10.1016/j.ijmedinf.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Uzuner O, Solti I, Cadag E. Extracting medication information from clinical text. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):514–518. doi: 10.1136/jamia.2010.003947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doan S, Bastarache L, Klimkowski S, Denny JC, Xu H. Integrating existing natural language processing tools for medication extraction from discharge summaries. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):528–531. doi: 10.1136/jamia.2010.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patrick J, Li M. High accuracy information extraction of medication information from clinical notes: 2009 i2b2 medication extraction challenge. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):524–527. doi: 10.1136/jamia.2010.003939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Z, Liu F, Antieau L, Cao Y, Yu H. Lancet: a high precision medication event extraction system for clinical text. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):563–567. doi: 10.1136/jamia.2010.004077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tikk D, Solt I. Improving textual medication extraction using combined conditional random fields and rule-based systems. J Am Med Inform Assoc. 2010 Sep-Oct;17(5):540–544. doi: 10.1136/jamia.2010.004119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medicine NLo RxNorm. 2009. http://www.nlm.nih.gov/research/umls/rxnorm/
- 11.Levin MA, Krol M, Doshi AM, Reich DL. Extraction and mapping of drug names from free text to a standardized nomenclature. AMIA Annu Symp Proc. 2007:438–442. [PMC free article] [PubMed] [Google Scholar]
- 12.Pustejovsky J, Castaño J, Ingria R, et al. TimeML: Robust Specification of Event and Temporal Expressions in Text; Fifth International Workshop on Computational Semantics; 2003. [Google Scholar]
- 13.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J Am Med Inform Assoc. 2010 Jan-Feb;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Birdwell KA, Grady B, Choi L, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet Genomics. 2012 Jan;22(1):32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xu H, Jiang M, Oetjens M, et al. Facilitating pharmacogenetic studies using electronic health records and natural-language processing: a case study of warfarin. J Am Med Inform Assoc. 2011 Jul-Aug;18(4):387–391. doi: 10.1136/amiajnl-2011-000208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ferrucci D, Lally A. UIMA: an architectural approach to unstructured information processing in the corporate research environment. Nat Langs Eng. 2004;10(3–4):327–348. [Google Scholar]
- 17.Jaccard P. The distribution of the flora in the alpine zone. New Phytologist. 11(2):37–50. [Google Scholar]