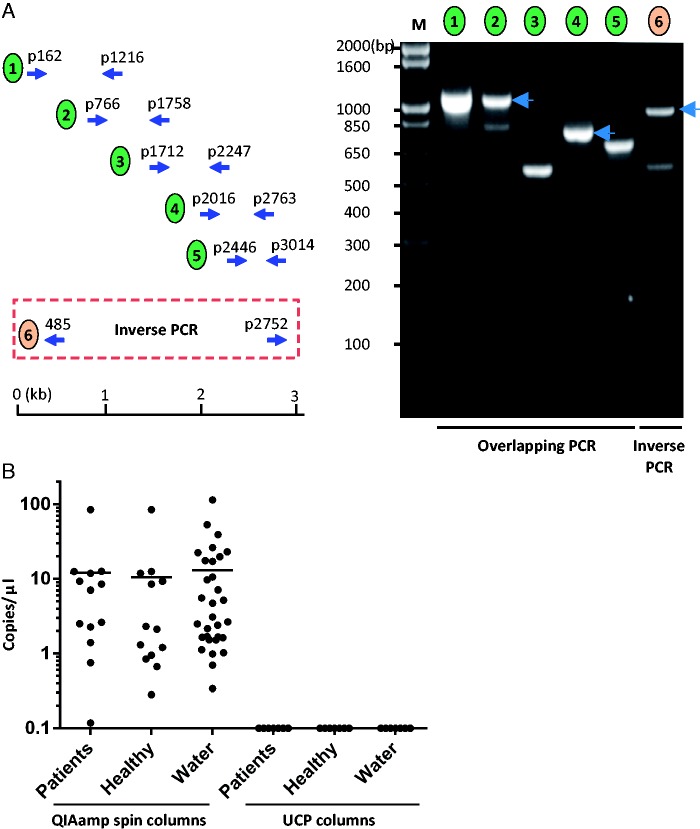
Fig. 1.—
Confirmation of contaminated columns as origin of CHIV14 DNA by qPCR. (A) Verification of CHIV14 genome assembled from the Illumina deep-sequencing data by overlapping PCR and inverted PCR. Five sets of overlapping primer pairs and one set of inverted primer pair were designed and used to amplify overlapping DNA fragments. (Left) Schematic diagram of the positions of primer pairs for the overlapping PCR and inverted PCR. (Right) Amplification overlapping viral DNA fragments. The numbers above indicate the primer pair used for the PCR as illustrated on the left. The numbers on the left indicate the molecular weight in base pairs. (B) Scatterplot showing copy number of CHIV14 per microliter of DNA extraction. DNA from patients (n = 13), healthy controls (n = 13), and water (n = 31) was extracted using QIAamp mini spin columns (QIAamp Viral RNA Mini kit; Qiagen). In parallel, seven DNA extractions for each specimen type (patients, healthy individuals, and water) were performed using the UCP columns. Each dot represents one specimen. Bars show the average copy numbers of the viral genome.