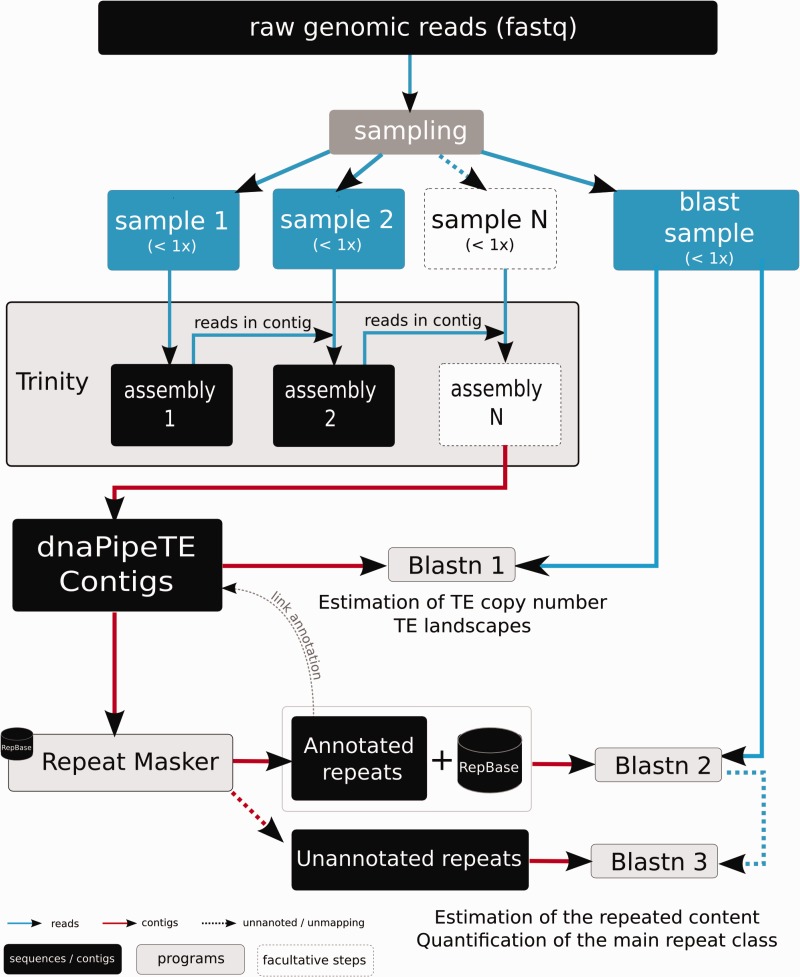
Fig. 1.—
Overview of the dnaPipeTE pipeline. First, genomic reads in FASTQ format are sampled. Then, assembly of repeats is performed using two or more iterations of Trinity. For each iteration, the previously assembled reads are added to the next sample to improve the repeat assembly. In the next step, assembled contigs are annotated using RepeatMasker. Finally, reads from the “BLAST sample” are blasted against all the contigs to estimate the relative abundance of each assembled repeat and to compute the TE landscape. In a second BLAST, the same sample is successively blasted against the annotated contigs joined to the Repbase library, then with the unannotated contigs in order to retrieve copies that would not have been assembled and to obtain a more global repeat content estimation. See text for additional details.