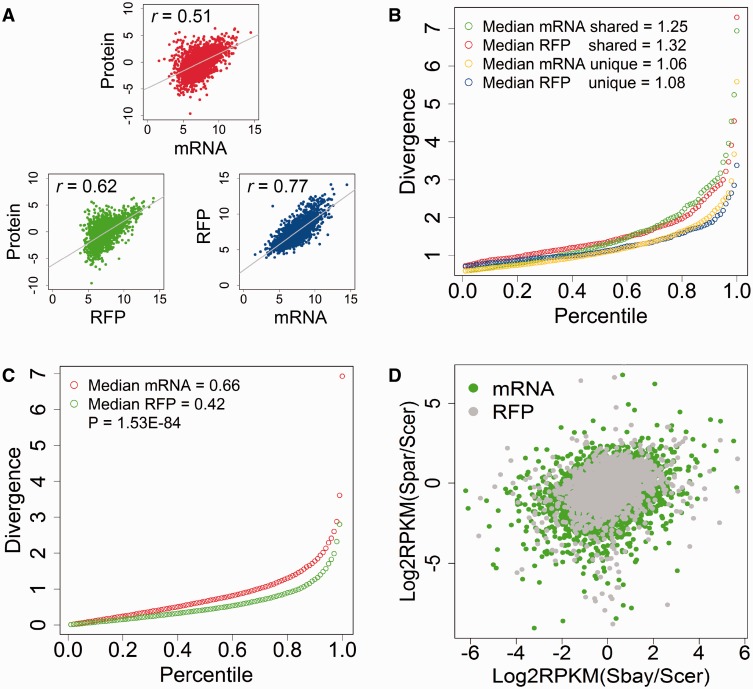
Fig. 2.—
Comparison of transcriptome, translatome, and proteome in yeast species. (A) Pearson correlation coefficient (r) between transcriptome (mRNA), translatome (RFP), and proteome (Protein) data in Saccharomyces cerevisiae. Reads per kilo per million (RPKM) for mRNA and RFP were calculated and compared with the normalized proteome data from Picotti et al. (2013). The line in each plot indicates the linear fitting with P values all essentially 0. (B) Comparison of transcription and translation for genes that had significant changes between the parental strains in mRNA (2,509 genes) and RFP (901 genes). The “shared” means that those genes had significant changes in both mRNA and RFP (559 genes), whereas the “unique” means that those genes had changes in either mRNA or RFP, but not both. Divergence was defined as |log2(fold change)| between the parental strains. The figure shows the quantile-normalized divergence for mRNA and RFP, and the number indicated the median divergence for the gene group. The shared genes have significantly higher divergence than the unique genes (WRT: P = 1.24E−10 for mRNA and P = 1.59E−11 for RFP). (C) The quantile-normalized divergence of transcription and translation for all genes. The transcription (median mRNA divergence = 0.66) has a significantly higher divergence than the translation (median RFP divergence = 0.42, WRT: P = 1.53E−84). (D) Comparison of the transcription and translation divergence between three yeast species, S. cerevisiae, S. bayanus, and S. paradoxus. The RPKM values of mRNA and RFP for each species were calculated and averaged between replicates. The log2-transformed RPKM ratios of transcription and translation were plotted. The plot for mRNA is more dispersed than RFP for the same species pairs. Significant more transcription than translation divergence was observed between S. cerevisiae and S. paradoxus (WRT: P < 2.22E−16, data were obtained from McManus et al. [2014]).