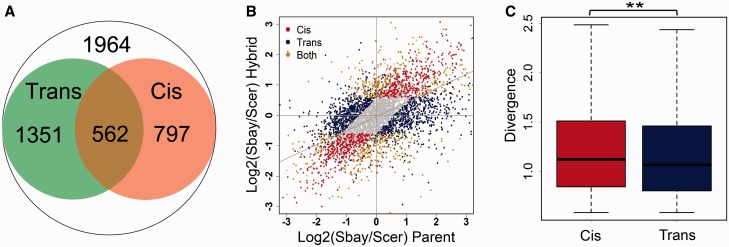
Fig. 6.—
The cis and trans effects on the evolution of TE. (A) TE was defined by dividing the RFP reads with the mRNA reads for each gene. The cis effect was obtained by comparing the TE differences between alleles in the hybrid strain (FDR ≤ 0.05 and fold change ≥1.5, 1,359 genes in total). The trans effect was defined by measuring the TE ratio changes of alleles between the parental strains and the hybrid (1,913 genes). The overlapping genes from the two categories have both the cis and trans effects in their TE regulation (562 genes). (B) Scatter plot for the cis, trans, and CT genes in TE regulation. The gray points show genes without allelic TE divergence (FDR > 0.05 or fold change <1.5), and the gray diagonal line represents the linear fitting using all the data. (C) Boxplot for the TE divergence between the parental strains for the cis and trans genes. The divergence of the cis effect genes was slightly higher than that of the trans genes (**, WRT: P < 0.01).