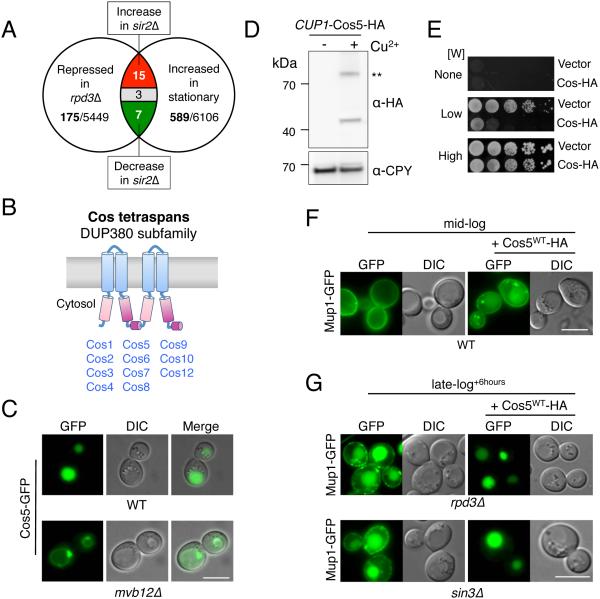
Figure 2. COS genes encode MVB sorting accelerators.
A) Venn diagram summarizing microarray mRNA expression data mined for genes that increase expression upon entry towards stationary phase and upon SIR2 deletion, and decrease expression upon loss of RPD3.
B) A cartoon of the predicted secondary structure for Cos proteins, which contain 4 transmembrane-spanning domains (TMDs). Pink and purple regions indicate internal regions sharing primary structural homology.
C) Localization of Cos5-GFP in wild-type cells and in mvb12Δ cells, which are defective for MVB sorting.
D) Immunoblot of WT cells carrying a low-copy plasmid encoding C-terminally HA-tagged Cos5 expressed from the inducible CUP1 promoter. Cells were gown in either 50 µM copper chelator (BCSA: bathocuproinedisulfonate) or 50 µM CuCl2 (− + Cu2+, respectively) prior to lysis, SDS-PAGE, and immunoblotting. A dimeric form of Cos5-HA is indicated (**).
E) Wild-type cells carrying CUP1-Cos5-HA or vector plasmid were serially diluted and grown on SD plates containing 50 µM CuCl2 and either no Trp (None), 4 µg/ml Trp (Low), or 20 µg/ ml Trp (High).
F) Localization of Mup1-GFP in wild-type cells grown to mid-log phase in SD-Met media containing 50 µM CuCl2. Cells were carrying vector plasmid or the CUP1-Cos5-HA plasmid.
G) Localization of Mup1-GFP in rpd3Δ and sin3Δ null cells carrying vector alone or the CUP1-Cos5-HA plasmid that were grown to late-log phase + 6 hours in SD-Met media containing 50 µM CuCl2. Bar = 5 µm.