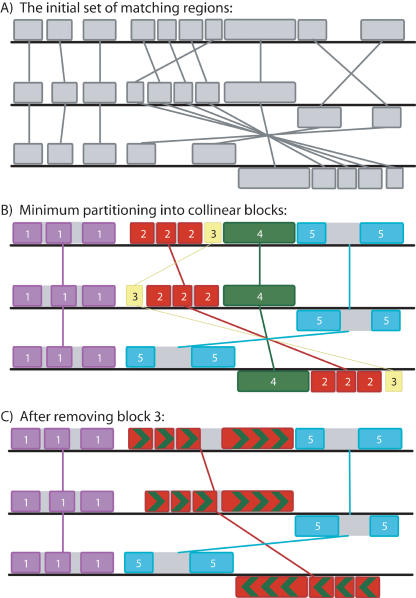
Figure 1.
A pictorial representation of greedy breakpoint elimination in three genomes. (A) The algorithm begins with the initial set of matching regions (multi-MUMs) represented as connected blocks. Blocks below a genome's center line are inverted relative to the reference sequence. (B) The matches are partitioned into a minimum set of collinear blocks. Each sequence of identically colored blocks represents a collinear set of matching regions. One connecting line is drawn per collinear block. Block 3 (yellow) has a low weight relative to other collinear blocks. (C) As low-weight collinear blocks are removed, adjacent collinear blocks coalesce into a single block, potentially eliminating one or more breakpoints. Gray regions within collinear blocks are targeted by recursive anchoring.