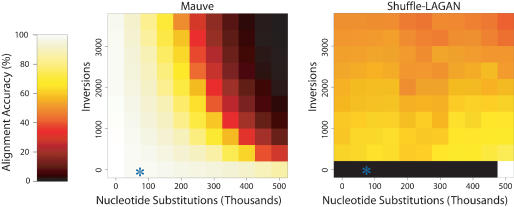
Figure 4.
The performance of Mauve (left) and Shuffle-LAGAN (right) when aligning two sequences evolved with increasing amounts of nucleotide substitution and inversions. Mauve is clearly more accurate than Shuffle-LAGAN at lower substitution rates. Shuffle-LAGAN version 1.2 did not complete some alignments without rearrangements, resulting in black entries. The observed substitution and inversion rate in the enterobacteria is denoted by an asterisk (*).