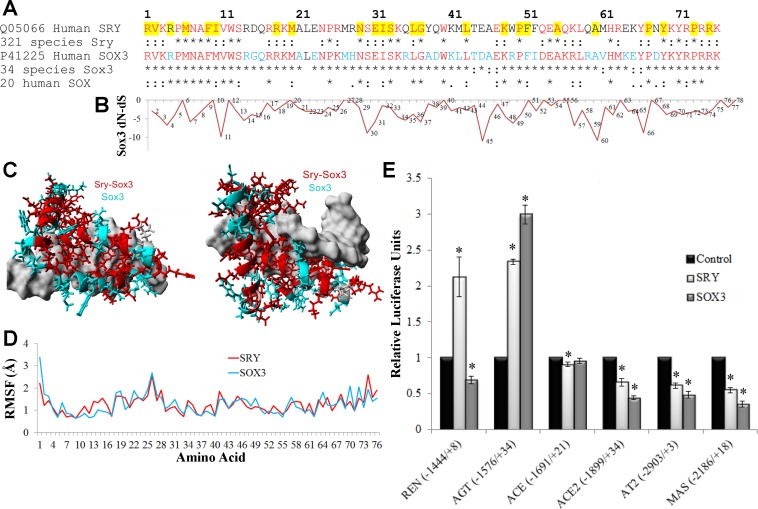
Fig. 1.
Functional conservation in SRY and SOX3. A: sequence alignment of the high-mobility group (HMG) box of human SOX3 and SRY with conservation shown below each in multiple species. Those sites marked with an asterisk (*) are 100% conserved in all species and those with a colon (:) are functionally conserved. Amino acids in red are conserved in all SRY and SOX3, while those in cyan are conserved in SOX3 only. Amino acids highlighted in yellow are those in which mutations in SRY are associated with sex reversal (50). B: normalized values for each amino acid's (x-axis) nonsynonymous rate (dN) minus the synonymous rate (dS) for the 34 sequences of Sox3. Values more negative suggest codon selection. C: known structure of SRY bound to DNA (pdb file 1j46) with the amino acids conserved among all SRY and SOX3 sequences shown in red and those conserved in SOX3 but not SRY shown in cyan. DNA is shown in gray. D: root-mean squared fluctuation (RMSF) for each amino acid over a 3.25 nanosecond molecular dynamic simulation for the known structure of SRY (red) or the modeled SOX3 (cyan) indicate similarities of the 2 proteins. E: the pEF1 expression vectors for either human SRY (light gray) or SOX3 (darker gray) were transfected into CHO cells along with a pGL3 vector containing the human promoter (shown on the bottom) of various RAS genes driving the production of luciferase. Each sample was normalized to an empty vector control of pEF1 (black). Error bars are shown as the SE. Each sample is compared with the control for each promoter. *Significant differences (P ≤ 0.05).