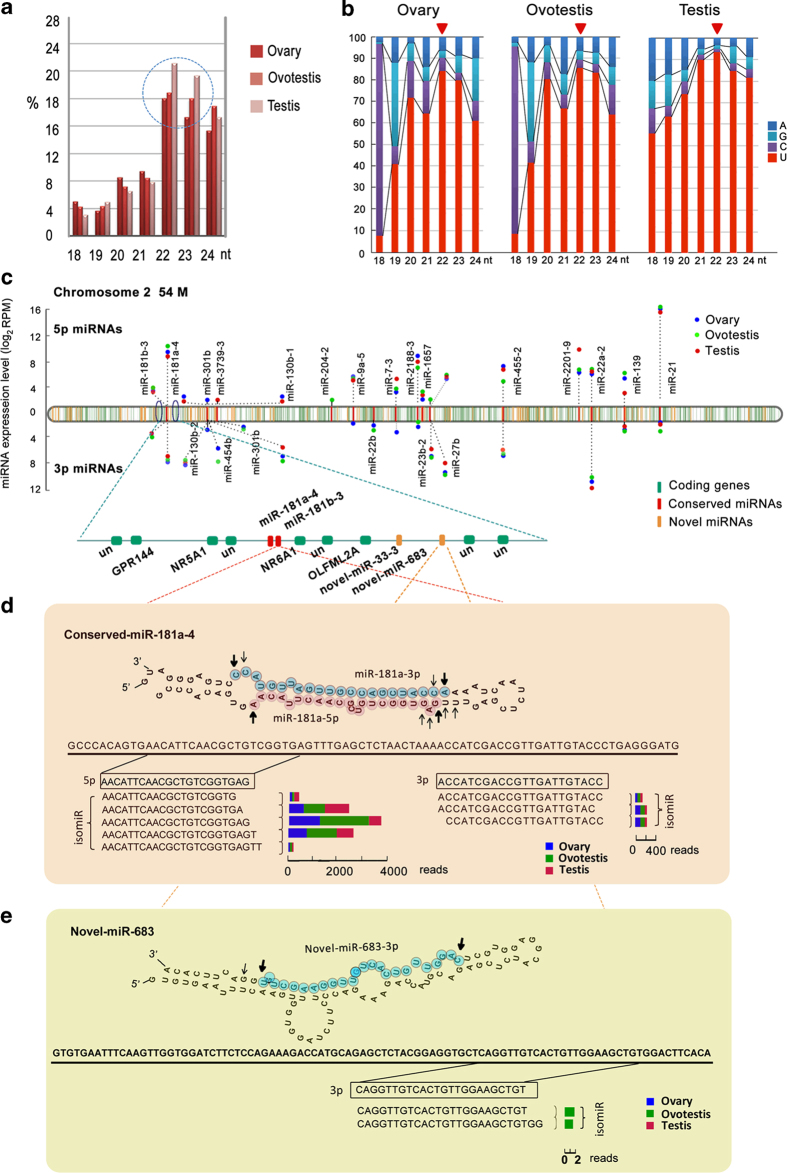
Figure 3.
Structure features of miRNAs during gonad reversal. (a) Length distributions of the miRNAs cloned from ovary, ovotestis and testis respectively, which exhibit characteristic sizes in 22-23 nt. The doted cycle indicates that the numbers of the miRNAs that range from 22 to 23 nucleotides are markedly raised in testis. (b) The preference distribution for uridine at the 5’ end position of different lengths (nt) of miRNAs in the ovary, ovotestis and testis. The miRNAs in the ovary and ovotestis show a remarkable loss in preference for uridine at the 5’ end position. (c) The chromosome 2 harbors 25 conserved miRNAs (red), 126 novel miRNAs (orange), in addition to 1754 protein-coding genes (green). Most of the miRNA precursors are located in intergenic regions. Over half of the stem loops can generate both 5’ and 3’ miRNAs from both arms. The conserved miRNAs are enriched in ovotestis. RPM, Reads Per Million reads. (d) The conserved miRNAs are mainly derived from both arms of a precursor (both 5’ and 3’). miR-181a-4 as an example for conserved miRNA biogenesis shows that differential processes at 5’ or 3’ end generate various isomiRs, which are differentially present during gonad reversal. (e) Novel miRNAs are mainly derived from one (either 5’ or 3’) arm of a precursor. Novel-miR-683 is used as an example for novel miRNA biogenesis. Multiple isomiRs are processed, most of which are present in one type of gonads. The length of rectangles indicates the reads of the isomiR in different types of gonads (ovary, blue; ovotestis, green; testis, red). The dominant cleavage sites are shown by arrows. Bold arrows show the most abundant isomiR cleavage sites, whereas thin arrows indicate less abundant isomiR cleavage sites.