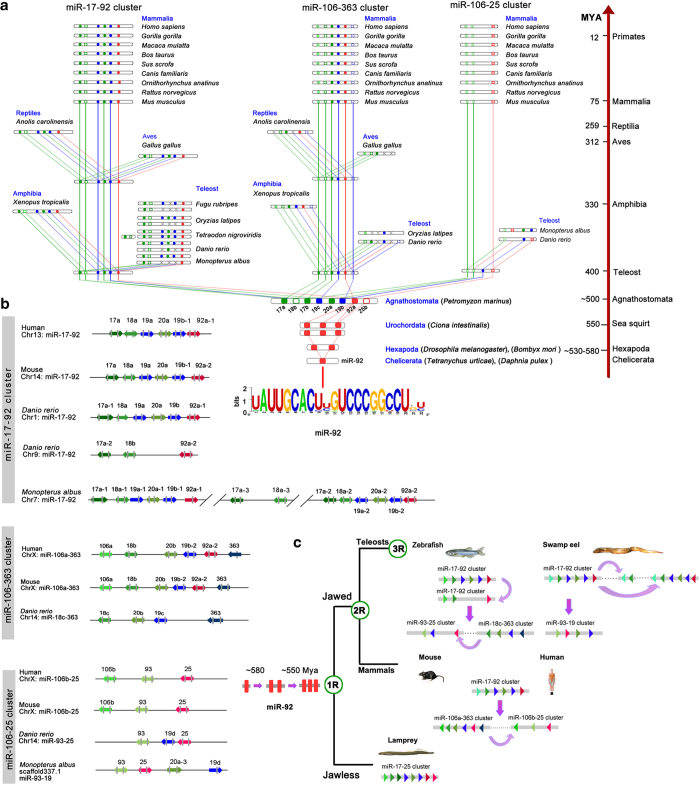
Figure 7.
Evolutionary trajectory of miR-17-92 cluster. (a) Phylogenetic analysis of the miR-17-92 cluster. Original miRNA cluster in lamprey has eight members including miR-17a, −18b, 17b, −19c, −20a, −19b, −92a and −25b. Jawed vertebrates have miR-17-92, miR-106-363 and miR-93-25 clusters. Both Tetranychus urticae and Daphnia pulex in Arthropoda have single copy miR-92. Both fruitfly (Drosophila melanogaster) and silkworm (Bombyx mori) have two linked copies (miR-92a/b). In sea squirt, miR-92 was duplicated again to generate two clusters, each with three members (miR-92a/b/c/d/e-1/e-2) on different chromosomes. Consensus sequence uAUUGCACugUCCCgGcCU of the miR-92 is indicated at the bottom on the panel. Divergence times are indicated on the right (MYA, million years ago). Green: members of the miR-17 family; blue: members of the miR-19 family; Red: members of the miR-25 family. (b) The genomic arrangements of miR-17-92 cluster and its paralogues in vertebrates. (c) An evolution model for miR-17-92. Original miR-17-92 cluster, which originated from miR-92, formed two clusters after 2nd whole genome duplication, one of which duplicated again within chromosome in mammals. In teleost fish, miR-17-92 was duplicated along with 3rd whole genome duplication. Duplicated cluster members could be lost, however, intra-chromosome duplications frequently occurred to compensate the miRNA loss. During evolution, deletions and rearrangements of miRNAs within the cluster also occurred.