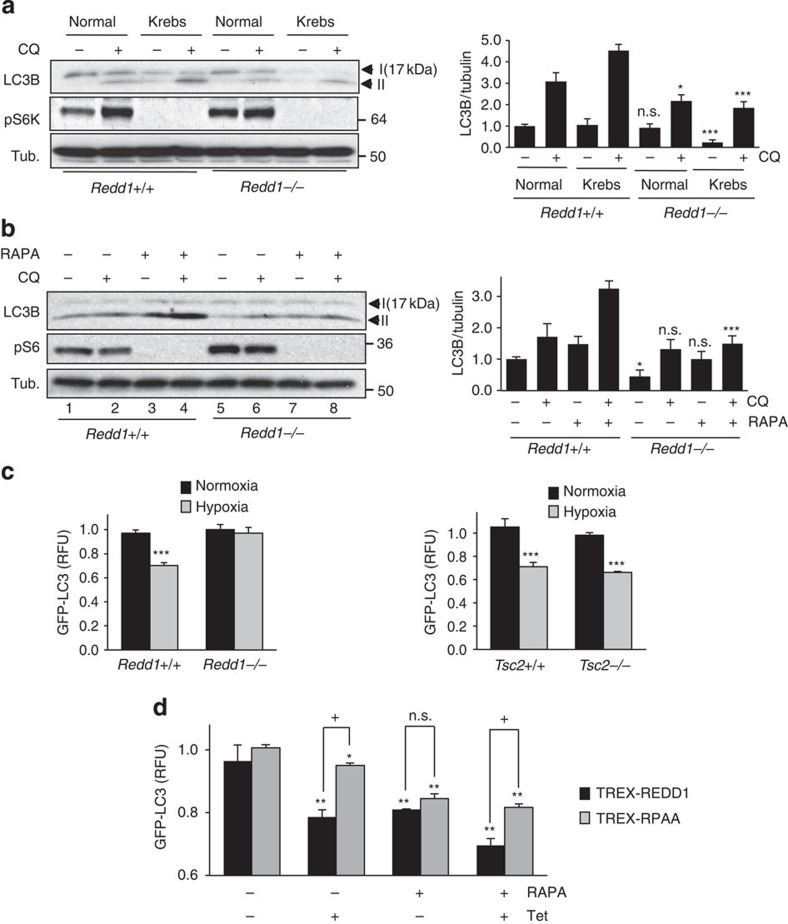
Figure 2. REDD1 mediates autophagy independent of mTORC1.
(a) Impaired starvation-induced autophagy in Redd1−/− cells despite suppressed mTORC1, assessed by the analysis of phosphorylated p70 S6 Kinase (pS6K, T389). Cells were cultured with Krebs media and treated in the absence or presence of CQ (30 μM, 1 h). At right, densitometry showing the mean values of three independent experiments and statistical comparison with respective wild-type lanes. (b) Rapamycin (100 nM, 4 h) fails to fully restore the defective autophagic flux in Redd1−/− cells, as assessed by LC3B processing in the absence or presence of CQ (30 μM, 4 h; compare lanes 4 and 8). Activity of mTORC1 was assessed via phosphorylated ribosomal protein S6 (pS6, S235/S236). Right, densitometry quantitation of three independent experiments as in a. (c) Failure of hypoxia (1% O2, 12 h) to induce autophagy in Redd1−/− cells (left) but not Tsc2−/− cells (right), assessed by degradation of GFP–LC3 measured using flow cytometry. (d) Both wild-type REDD1 and the mTORC1-inactive mutant REDD1-RPAA14 are sufficient to induce autophagy, assessed by GFP-LC3 levels using flow cytometry. Tetracycline (Tet)-inducible (TREX) cells were treated in the presence and absence of rapamycin (100 nM, 4 h) as a positive control. For c,d, shown are the means of two independent experiments, each performed as triplicate cultures; error bars, s.d. (*) show P-values compared with untreated control; (+) compares REDD1 with REDD1-RPAA. For all experiments: ***P<0.001, **P<0.01, *P<0.05; +P<0.05, by paired t-test.