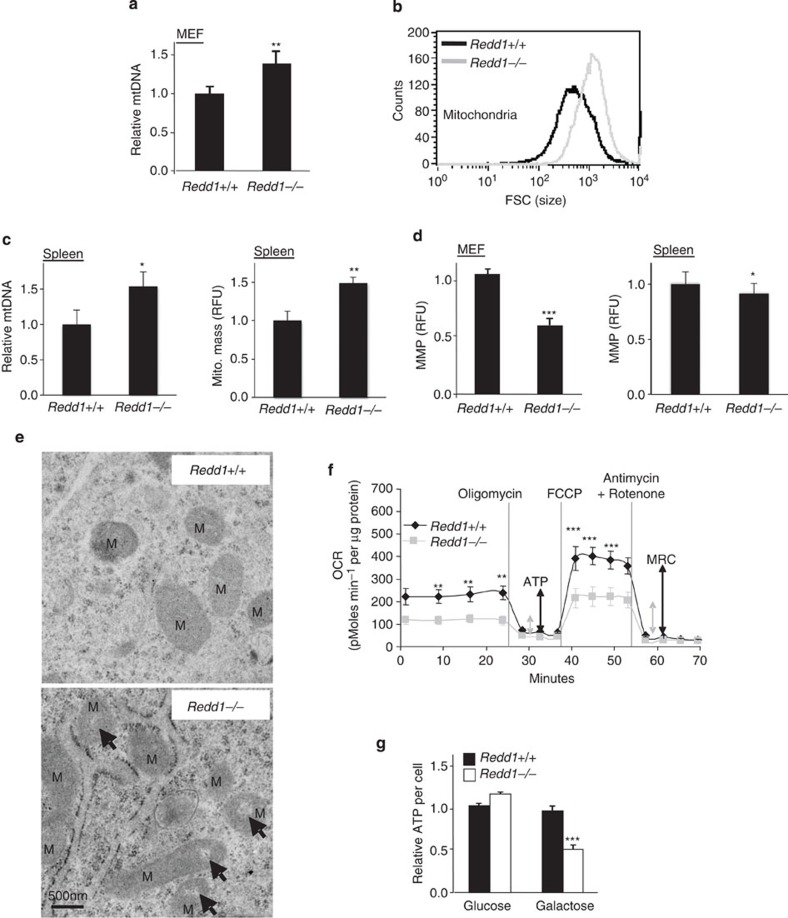
Figure 3. Loss of REDD1 results in accumulation of dysfunctional mitochondria.
Redd1−/− cells exhibit (a) increased mitochondrial (mt) number, evidenced by increased mitochondrial to nuclear (M/N) DNA ratio (COXII/β-globin) as measured by QRT–PCR. MEF, mouse embryonic fibroblast. PMEF, primary MEF. Shown are the mean from two independent experiments, each performed as triplicate cultures. (b) Increased mitochondrial size as determined using flow cytometry of cell-free, purified mitochondria. (c) Increased mitochondrial number (left) and mass (right) in Redd1−/− primary splenocytes from starved mice (24 h), assessed, respectively, by M/N DNA ratio (N=4 mice per genotype, analysed in duplicate) and by staining with Mitotracker Green (100 nM) and then flow cytometry analysis (N=3 mice per genotype, analysed in triplicate). (d) Reduced mitochondrial membrane potential (MMP) in Redd1−/− MEFs (left) and splenocytes (right) as assessed by staining with Tetramethylrhodamine, Ethyl Ester (TMRE, 100 nM) and then flow cytometry (N=2 mice per genotype, analysed in triplicate). (e) Dysmorphic mitochondria (M) in Redd1−/− cells as assessed using electron microscopy. Arrows indicate abnormal hypodense regions. (f) Decreased basal O2 consumption rate (OCR), mitochondrial ATP synthesis (ATP) and maximal respiratory capacity (MRC) in Redd1−/− cells, measured via Seahorse XF24. (g) Impaired oxidative phosphorylation in Redd1−/− MEFs, shown by a drop in ATP per cell on shift from glucose to galactose-containing media (48 h). All error bars show s.d. ***P<0.001, **P<0.01, by paired t-test (a,d,e,g,h).