Figure 4.
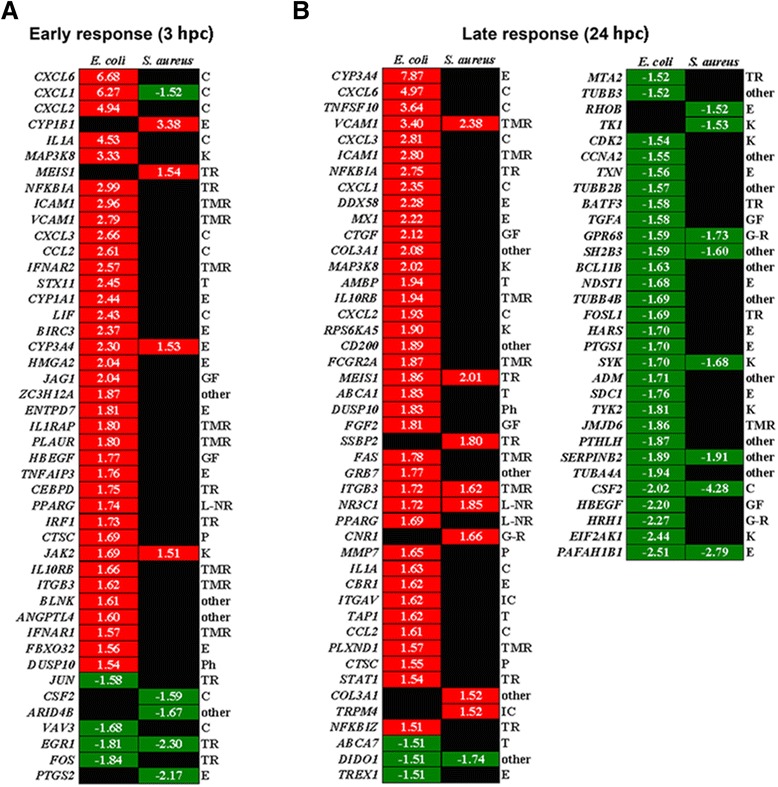
Differentially expressed genes associated with “inflammatory response” in PMEC after pathogen challenge. Heat maps show differentially expressed genes annotated by IPA and grouped according to their maximal altered mRNA concentrations as well as a function of challenge time (red, up-regulated; green, down-regulated; fold changes are given inside the boxes). (A) More genes were affected at 3 hpc (early response) with E. coli (40 genes) than with S. aureus (9 genes). (B) The majority of differentially expressed genes of PMEC was also involved in late response (24 hpc) to challenge with E. coli (70 genes) than to challenge with S. aureus (17 genes). Gene functions according to the IPA annotation are given to the right. The affected inflammatory response genes encoding cytokines (C), enzymes (E), kinases (K), transcription regulators (TR), transmembrane receptors (TMR), transporter (T), growth factors (GF), ligand-dependent nuclear receptors (L-NR), peptidases (P), phosphatases (Ph), G-protein coupled receptors (G-R) and ion channel proteins (IC).
