Figure 1.
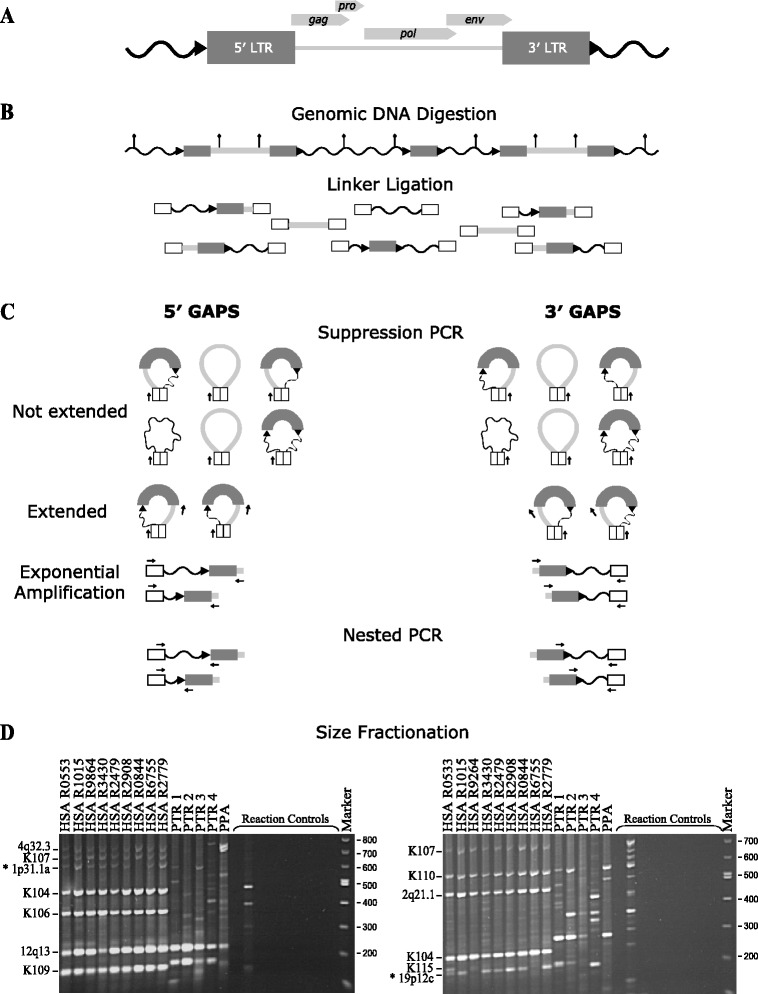
Schematic representation of GAPS and size fractionation of amplicons from humans and chimpanzees. (A) HERV-K(HML-2) provirus containing viral genic regions gag, pro, pol and env bordered by two LTRs, direct repeats (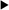 ) and unique flanking DNA (
) and unique flanking DNA ( ). (B) Linkered genomic DNA library generation. Restriction sites (
). (B) Linkered genomic DNA library generation. Restriction sites ( ) within a representative section of genomic DNA that contains two proviruses and a solo LTR is shown at the top. Following digestion annealed linkers (white boxes) are ligated to genomic DNA restriction fragments, which then serve as templates for PCR. (C) GAPS PCR amplifications. 5′ GAPS targets the 5′ end, and 3′ GAPS the 3′ end of proviral sequences. Arrows (
) within a representative section of genomic DNA that contains two proviruses and a solo LTR is shown at the top. Following digestion annealed linkers (white boxes) are ligated to genomic DNA restriction fragments, which then serve as templates for PCR. (C) GAPS PCR amplifications. 5′ GAPS targets the 5′ end, and 3′ GAPS the 3′ end of proviral sequences. Arrows ( ) indicate primer binding sites. Following suppression PCR melting, self-complementary linkers form duplexes creating a population of stem-looped DNA molecules. Extension from the linker primer is prevented as primer annealing is blocked by linker duplex formation at the base of the stem. Molecules containing the binding site for the target primer of choice (GAG or ENV) are extended and become templates for exponential amplification. Nested PCR is performed to enrich for sequences containing a LTR and reduce amplicon length. (D) Size fractionation by gel electrophoresis. 5′ GAPS on left and 3′ GAPS on right. Genomes include nine humans (HSA) and four chimpanzees (PTR). Lane 14 (PPA) is a positive control utilising bonobo DNA. The remaining controls were for construction of linker-genomic DNA fragments and were set up using the same genomic DNA sample as Lane 9. Library controls (left to right): No genomic DNA, no restriction enzyme, no linker ligated genomic DNA, no ligase and no linker. The final four controls were PCR negatives with DNA omitted. Numbers on the left are DNA size marker fragments (bp). Loci retrieved using GAPS are indicated on the right. New polymorphic proviruses discovered using GAPS (*). Refer to Additional file 1 for the un-cropped gel image.
) indicate primer binding sites. Following suppression PCR melting, self-complementary linkers form duplexes creating a population of stem-looped DNA molecules. Extension from the linker primer is prevented as primer annealing is blocked by linker duplex formation at the base of the stem. Molecules containing the binding site for the target primer of choice (GAG or ENV) are extended and become templates for exponential amplification. Nested PCR is performed to enrich for sequences containing a LTR and reduce amplicon length. (D) Size fractionation by gel electrophoresis. 5′ GAPS on left and 3′ GAPS on right. Genomes include nine humans (HSA) and four chimpanzees (PTR). Lane 14 (PPA) is a positive control utilising bonobo DNA. The remaining controls were for construction of linker-genomic DNA fragments and were set up using the same genomic DNA sample as Lane 9. Library controls (left to right): No genomic DNA, no restriction enzyme, no linker ligated genomic DNA, no ligase and no linker. The final four controls were PCR negatives with DNA omitted. Numbers on the left are DNA size marker fragments (bp). Loci retrieved using GAPS are indicated on the right. New polymorphic proviruses discovered using GAPS (*). Refer to Additional file 1 for the un-cropped gel image.
