Figure 3.
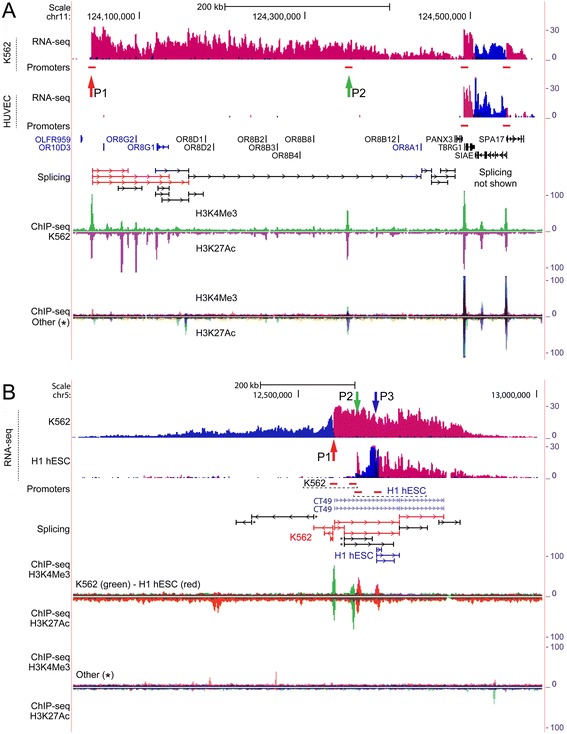
Complex regulation by ERVs is applicable also to annotated loci. (A) ERV9-LTR12 regulation of multiple olfactory gene-containing locus. Chimeric transcription from an ERV9-LTR repeat (P1, red vertical arrow) is associated with major activation of the olfactory locus, including (chimeric) splice junctions that span large distances in the locus. The RNA-seq coverage suggests that olfactory genes are activated strand-specifically (genes marked in blue are located on the positive strand) and include splicing into OR8G1. ChIP-seq shows that the position of the ERV9-LTR12 repeat coincides with a major H3K4Me3-H3K27Ac promoter marker in K562 cells. The role of a MIR in P2 is described in the main text. *Overlay view of ChIP-seq from Gm12878, H1 hESC, HSMM, HUVEC, NHEK and NHLF cell lines from ENCODE/Regulation. (B) Concurrent annotated and unannotated transcription of the CT49 locus. RNA-seq chimeric splice junctions show differential isoform expression of CT49 in H1 hESC (blue splice junctions) and K562 (red splice junctions). Transcription of CT49 is associated with unannotated transcription in the opposite direction in both cell lines. The positions P1-P3 coincide with ChIP-seq regulatory motifs in K562 (P1 and P2) and H1 hESC (P2 and P3), exclusively, showing that repeats present at these positions are major regulators of the CT49 locus (close-up views are shown in Figure 4). *Overlay view of ChIP-seq from Gm12878, HSMM, HUVEC, NHEK and NHLF cell lines from ENCODE/Regulation. Note that in this subfigure, P2 (green vertical arrow) marks two adjacently positioned promoter regions (horizontal red bars, not drawn to scale), i.e. one from both cell lines. Non-chimeric splice junctions marked with a star (*) are from K562, while the remaining black splice junctions are from either cell line. Please refer to Figure 1 legend for detailed description of data presentation.
