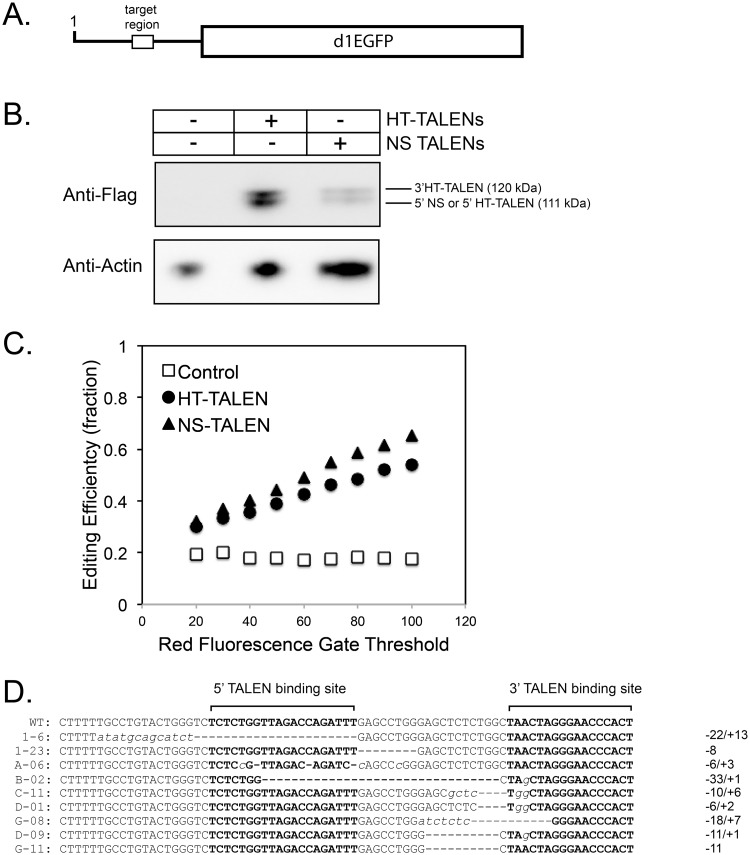
Fig 3. HT-TALEN and NS-TALEN targeting of an HIV-1 LTR in cell culture.
A. Schematic diagram of DNA GFP reporter to be targeted by HT-TALENs and NS-TALENs. The target DNA contains the 5’ LTR of HIV-1 fused upstream of the coding region of d1EGFP. B. Western blot analysis of HeLa-tat-III/LTR/d1EGFP cells transfected with either the HT-TALEN pair or NS-TALEN pair. The blot was probed with anti-Flag and anti-Actin as a loading control. C. Dose-response plot based on quantification of flow cytometry analysis of GFP reporter expression. Transiently transfected HeLa-tat-III/LTR/d1EGFP samples were analyzed for GFP and mCherry expression. Cells with mCherry contained the transfected plasmids. Cells containing the functional HIV-1 LTR fused d1EGFP reporter expressed GFP. Samples were done in triplicate. Those samples not expressing GFP, only mCherry were compared. Standard deviations from triplicate samples are smaller than the symbols and not shown. Statistically significant differences between slopes for TALEN treatment and control indicated is by a * (p<0.000001); NS-TALEN and HT-TALENs were not significantly different (p<0.08). D. Sequences of genomic clones containing mutated target regions. Upper-case bolded font indicates designated 5’ TALE and 3’ TALE binding sites. Inserted nucleotides are in lower-case italicized font. A deletion is represented by dashes. Lengths of the insertions (+) and deletions (-) are at the right of each sequence.