Figure 2.
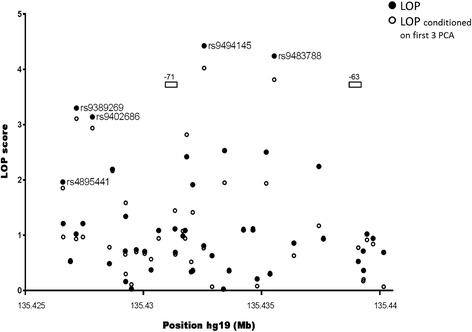
African-specific association with HbF at HMIP-2B in Tanzanian SCD patients. Association analysis was performed with the same individuals as in Figure 1, but 15 patients with Eurasian-type high-HbF haplotypes (‘A – B’, carrying the ancestry-informative allele rs1376090-C) were excluded. Data for 54 markers imputed from 1000 Genomes YRI (Yoruba, Ibadan, Nigeria) sequence were added to the analysis. While the exclusion of selected individuals resulted in a weaker overall association signal, the potential to map African-specific variants at higher resolution was considered more important at this stage. Association scores are shown; unconditioned (black dots) and conditioned on the first three principal components derived from genome-wide SNP data (open circles) [25]. Shown also is the location of the conserved MYB upstream regulatory elements −71 and −63 [13].
