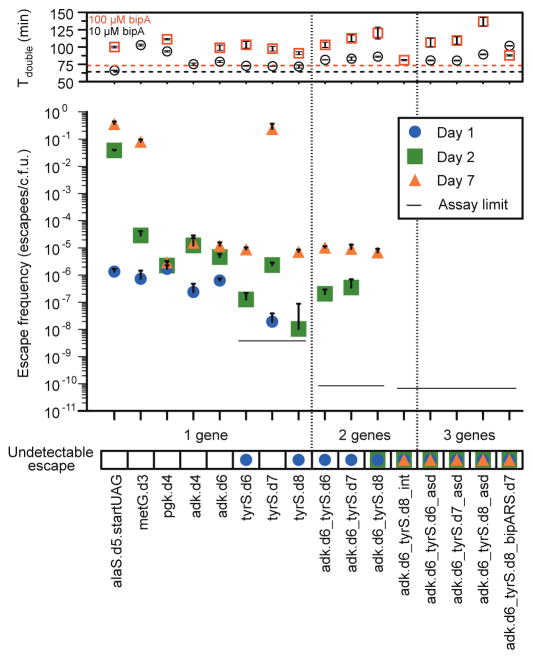
Figure 2. Escape frequencies and doubling times of auxotrophic strains.
Escape frequencies for engineered auxotrophic strains calculated as colonies observed per colony forming unit (c.f.u.) plated over 3 technical replicates on solid media lacking arabinose and bipA. Assay limit is calculated as 1/(total c.f.u. plated) for the most conservative detection limit of a cohort, with a single-enzyme auxotroph limit of 3.5 × 10−9 escapees/c.f.u., a double-enzyme auxotroph limit of 8.3 × 10−11 escapees/c.f.u. and a triple-enzyme auxotroph limit of 6.41 × 10−11 escapees/c.f.u. Positive error bars are standard error of the mean (s.e.m.) of the escape frequency over three technical replicates (Methods). The top panel presents the doubling times for each strain in the presence of 10 μM or 100 μM bipA, with the parental strain doubling times represented by the dashed horizontal lines. MetG.d3 growth was undetectable in 100 μM bipA. Positive and negative error bars are s.e.m.