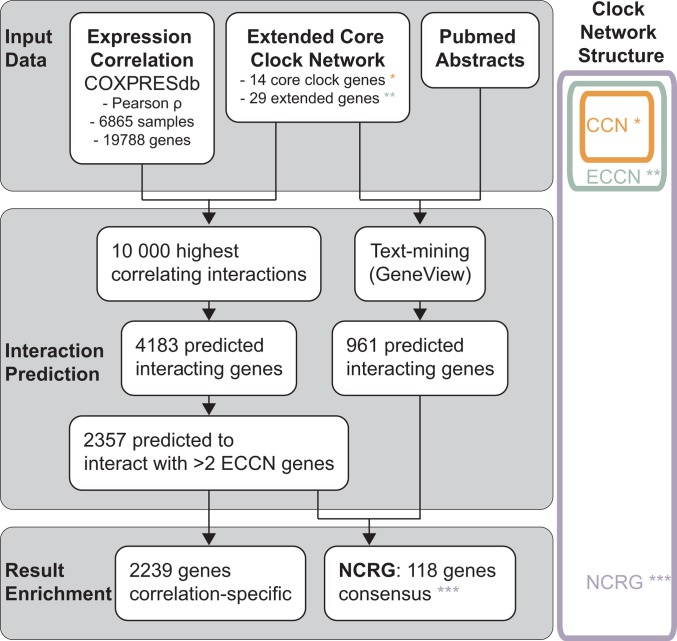
Fig 1. Work flow used to establish a network of circadian regulated genes (NCRG).
Two independent data types were used to predict genes which interact with the human core clock network (CCN, orange) and the extended core clock network (ECCN, green). Co-expression data was used to find sets of genes with strongest (anti-) correlating expression with the 43 ECCN genes across a large number of independent experiments. A total of 2357 genes were found to interact with more than 2 ECCN genes. The GeneView text-mining pipeline was used to analyse published knowledge (approximately 22 million citations) about interacting genes. A total of 961 text-mining-predicted genes were found. The intersection of both methodologies resulted in 118 new genes, which together with the ECCN form a new network of circadian regulated genes (NCRG, purple).