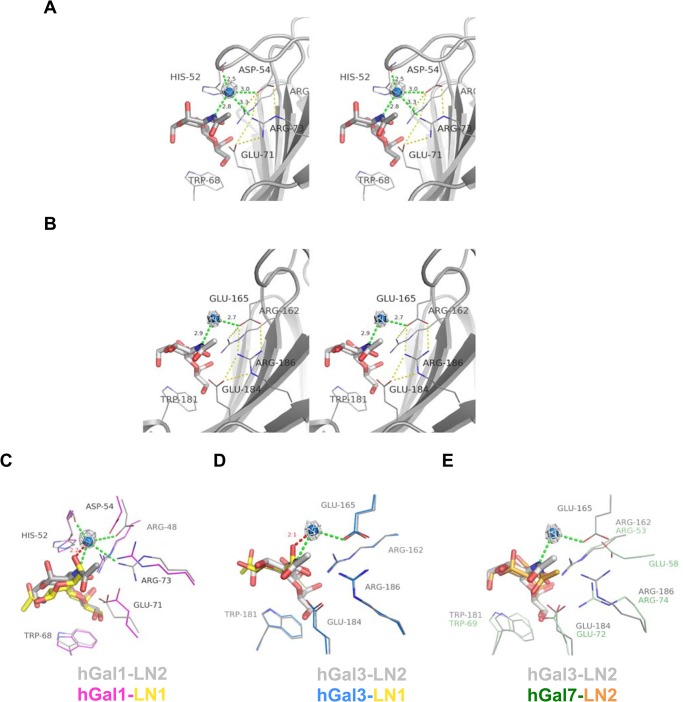
Fig 3. Water-mediated interactions of hGal1 and hGal3 with the LN2 molecules.
(A and B) Stereoview of LN2 molecule bound in the carbohydrate-recognition site of hGal1 (PDB ID: 1W6P) and hGal3-CRD (1KJL), respectively. LN2 ligand is depicted as gray stick model. The water (blue sphere) is coordinated by the H-bonds (green dashes) from N2 atom of LN2 and amino acid residues of galectins. 2F o -F c omit electron density (gray mesh) of the water molecules are highlighted and contoured at 1σ. Unique salt bridge network of hGal1 and hGal3-CRD are indicated as yellow dashes. (C and D) Structural superposition of LN1 and LN2 complex structures from hGal1 (C) and hGal3-CRD (D). The C5-hydroxyl group of LN1 (yellow sticks) in hGal1 and hGal3-CRD complexes makes close contact with the coordinated water in LN2-hGal1 andLN2-hGal3-CRD complex with a distance of 2.2 and 2.1 Å, respectively. (E) Structure of hGal7 (shown in color green) in complex with LN2 (orange) is superimposed with LN2-hGal3-CRD complex structure (all in gray). Most LN2-contacting residues in hGal7 (such as Arg53hGal7, Trp69hGal7, Glu72hGal7 and Arg74hGal7) are well superimposed with those of hGal3, except the Glu58hGal7 residue. Location of Glu58hGal7 is distinctive from Glu165hGal3/ Asp54hGal1 and distance between Glu58hGal7 and N2 atom of LN2 is too far for them to coordinate a water molecule in between.