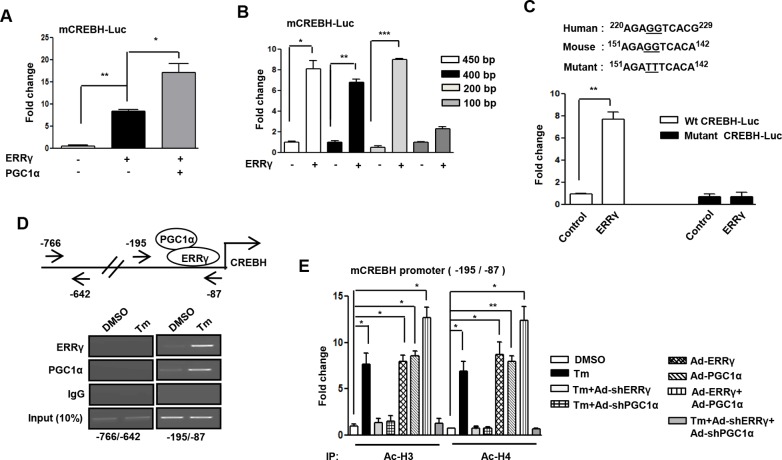
Fig 3. ERRγ regulates activation of CREBH gene promoter.
(A), PGC1α-dependent activation of the mouse CREBH promoter by ERRγ. Transient transfection was performed in 293T cells with the indicated plasmid DNAs. (B), Deletion constructs of the CREBH promoter demonstrate the ERRγ binding site in 293T cells. Transient transfection was performed in 293T cells with the indicated plasmid DNAs. (C), ERRE-dependent activation of the CREBH promoter in 293T cells. 293T cells were transfected with the wild-type or ERRE-mutant CREBH promoter along with ERRγ plasmid DNAs. (D), ChIP assay shows the binding of ERRγ and PGC1α to the endogenous CREBH promoter by semiquantitative PCR. AML12 cells were treated with DMSO or Tm (5μg/mL) for 12 hr. After completion of the treatment, chromatin fragments were prepared and immunoprecipitated with ERRγ, PGC1α, or IgG control antibodies. DNA fragments covering –766 to –642 and –195 to –87 elements on the CREBH promoter were PCR amplified. 10% of the soluble chromatin was used as input. (E), ChIP assay for detection of histone acetylation at the ERRγ/PGC1α binding site under the indicated conditions in AML12 cells. Chromatin fragments were prepared and immunoprecipitated with Acetyl-Histone 3 and Acetyl-Histone 4 antibodies. DNA fragments covering –195 to –87 element on the CREBH promoter were qPCR-amplified as described in the ‘Materials and Methods’ section. Data are representative of three independently performed experiments and shown as mean±SD; *P<0.05, **P<0.005, and ***, p<0.0005 using Student’s t-test.