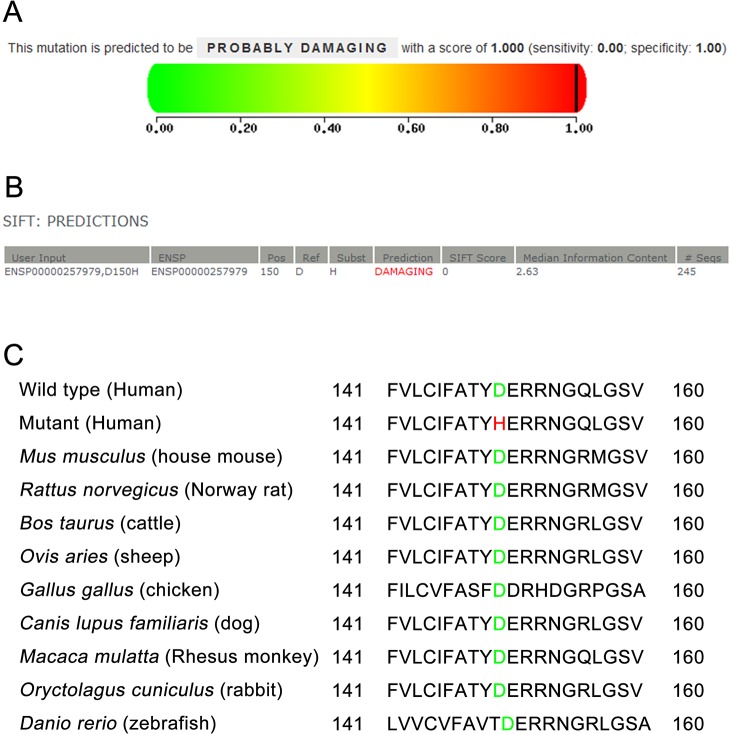
Fig 5. Bioinformatics analysis of the p.D150H mutation and multiple sequence alignment.
(A) The mutation was predicted to be probably damaging with a score of 1.00 by PolyPhen-2, which meant it may have deleterious effect on the structure and function of the protein. (B) the score and media information content from SIFT was 0.00 and 2.63 respectively, predicting the effect of this amino acid substitution on protein function was damaging. (C) The result of a multiple sequence alignment from various species showed that the aspartic acid at position 150 of AQP0 is highly conserved (marked in green, the mutation in red).