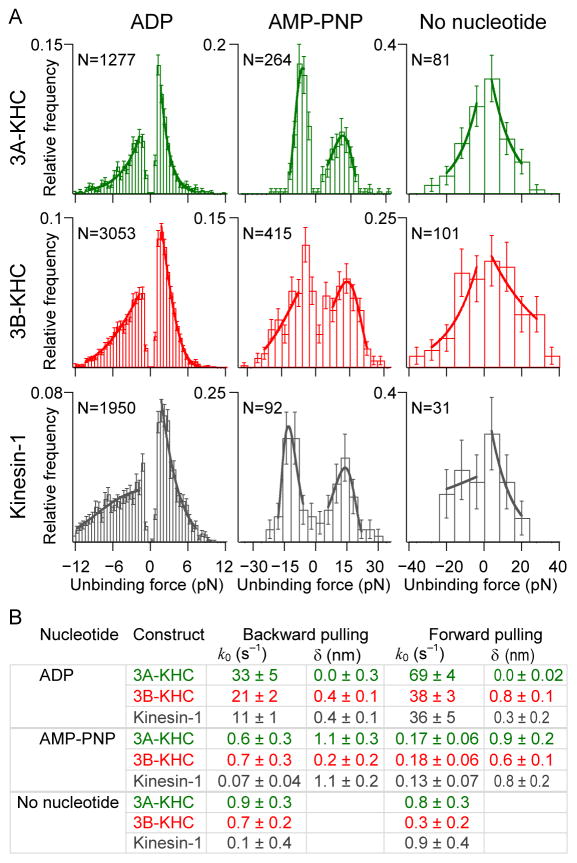
Figure 5. Unbinding force measurements.
(A) Unbinding force histograms for 3A-KHC, 3B-KHC and kinesin-1 (rows) in 1 mM ADP, 0.5 mM AMP-PNP, or no nucleotide (columns). Negative unbinding forces correspond to pulling kinesin toward the microtubule minus-end (the hindering load direction); positive forces are toward the plus-end (assisting load direction). Loading rates: 100 pN s−1 for ADP; 10 pN s−1 for AMP-PNP and no nucleotide (apyrase). Solid lines represent fits to the data under each curve for each pulling direction. Bins with few counts were excluded as well as data at low forces due to the possibility of missed events.
(B) Fit parameters (± s.e. of fit), are shown for each construct and experimental condition, using the parameters indicated in Eq. (1). k0 is the unloaded off-rate and δ is the characteristic distance parameter. For microtubule dissociation rates during processive stepping, see also Figure S4.