Abstract
The mycotoxin aflatoxin B1 (AFB1) may initiate cancer by causing oxidatively damaged DNA, specifically by causing 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxodG) lesions. Base excision repair removes these lesions, with 8-oxoguanine glycosylase (OGG1) being the rate-limiting enzyme. The aim of this study was to determine the effect of ogg1 deficiency on AFB1-induced oxidatively damaged DNA and tumourigenesis. Female wild-type, heterozygous and homozygous ogg1 null mice were given a single dose of 50mg/kg AFB1 or 40 µl dimethyl sulfoxide (DMSO) ip. Neither ogg1 genotype nor AFB1 treatment affected levels of oxidised guanine in lung or liver 2h post-treatment. AFB1-treated ogg1 null mice showed exacerbated weight loss and mortality relative to DMSO-treated ogg1 null mice, but AFB1 treatment did not significantly increase lung or liver tumour incidence compared with controls, regardless of ogg1 genotype. Suspect lung masses from three of the AFB1-treated mice were adenomas, and masses from two of the mice were osteosarcomas. No osteosarcomas were observed in DMSO-treated mice. All liver masses from AFB1-treated mice were adenomas, and one also contained a hepatocellular carcinoma. In DNA from the lung tumours, the K-ras mutation pattern was inconsistent with initiation by AFB1. In conclusion, ogg1 status did not have a significant effect on AFB1-induced oxidatively damaged DNA or tumourigenesis, but deletion of one or both alleles of ogg1 did increase susceptibility to other aspects of AFB1 toxicity.
Introduction
Aflatoxin B1 (AFB1) is a known human liver carcinogen and potential human lung carcinogen (1–3). Produced by species of Aspergillus, AFB1 contaminates food crops in humid and hot conditions. One mechanism by which AFB1 is believed to cause cancer requires biotransformation of AFB1 into a reactive epoxide that binds to guanine residues in DNA, forming DNA adducts; if left unrepaired, these adducts can cause mutations that may lead to cancer (4,5). However, AFB1 has also been shown to cause oxidatively damaged DNA, which could potentially result in the same mutations caused by AFB1 alkylation, also leading to cancer (6,7).
The most abundant and commonly studied product of oxidatively damaged DNA is 8-oxo-7,8-dihydro-2’-deoxyguanosine (8-oxodG). 8-oxodG can be produced by the attack of the hydroxyl radical on C-8 of guanine, and is one of the most highly mutagenic DNA lesions (8,9). In vivo treatment with AFB1 causes an increase in 8-oxodG in mouse lung DNA (10,11), and 8-oxodG lesions can cause G to T transversions and G to A transitions (12), which are consistent with K-ras mutations found in AFB1-induced mouse lung tumours (13,14). Specifically, when 8-oxodG is paired with adenine, it is able to mimick thymine functionally by adopting the syn conformation around the N-glycosidic bond, establishing the primary structural basis for 8-oxodG mutagenicity, resulting in G to T transversions (12).
Oxidatively damaged DNA is repaired primarily by base excision repair (BER). BER handles the largest number of cytotoxic and mutagenic base lesions, (12,15) and is the main mechanism for the removal of DNA lesions that cause minor helix distortions, such as 8-oxodG damage (16,17). 8-Oxoguanine glycosylase (OGG1) is the rate limiting enzyme in the BER pathway that specifically excises 8-oxoG lesions (9). Interestingly, acute exposure to a tumourigenic dose of AFB1 increases 8-oxodG levels in mouse lung, which corresponds with an increase in BER activity and an increase in OGG1 protein levels (11). As such, AFB1-induced oxidatively damaged DNA may be an important contributing process in AFB1-induced carcinogenesis. Mice deficient in ogg1 have been developed and allow for the ability to determine the consequences of 8-oxodG lesions specifically.
To date, the role of oxidatively damaged DNA in AFB1-induced tumourigenesis has not been directly assessed. In this study, we investigated the impact of deletion of just one or both ogg1 alleles on oxidatively damaged DNA and tumourigenesis following a single tumourigenic dose of AFB1.
Materials and methods
Materials
Chemicals were obtained as follows: AFB1, alkaline phosphatase, nuclease P1 from Sigma (St. Louis, MO, USA). Ultra-free-MC 10000 nominal molecular weight limit (NMWL) filter units from Millipore (Bedford, MA, USA). Ten percent neutral buffered formalin and histology cassettes from Fisher Scientific (Toronto, ON, Canada). All other chemicals were reagent grade and were obtained from common commercial suppliers.
Animals and housing conditions
Mice deficient in the ogg1 gene on a 129/C57Bl/6 genetic background were generated as described (18). ogg1 knockout mice were kindly provided by Dr. Christi A. Walter at the University of Texas Health Science Center at San Antonio (San Antonio, TX, USA) and maintained in the Queen’s University animal facility. The genotype of each mouse was determined by isolating DNA from a small portion of each mouse’s tail at weaning, using a Qiagen DNeasy Blood and Tissue kit (Toronto, ON, Canada) and a polymerase chain reaction (PCR) analysis was performed using a Qiagen multiplex PCR kit (Toronto, ON, Canada). The primer sequences have been published previously (19). The PCR conditions were 95°C for 15:00min, (94°C for 0:30min, 58oC for 1:30min, 72°C for 1:30min) repeated 30 times, 72oC for 10:00min, and 15°C until sample retrieval.
All animal experiments were conducted in accordance with institutional guidelines and the policies of the Canadian Council on Animal Care.
Measurement of oxidatively damaged DNA
Female wild-type mice (+/+) as well as those with a heterozygous (+/−) or homozygous (−/−) deficiency of the ogg1 gene, aged 7–10 weeks, were housed with a 12-h light/dark cycle and provided food and water ad libitum. Mice were treated with either 50mg/kg AFB1, which results in pulmonary tumourigenesis in AC3F1 mice and A/J mice (13,14) or with 40 µl dimethyl sulfoxide (DMSO) ip. One hundred minutes post-dosing, mice were given heparin ip (1.12mg/mouse in 0.1ml sterile saline) to aid in tissue perfusion. Two hours post AFB1 treatment, mice were killed by cervical dislocation. Lungs and livers were perfused with Tris–EDTA (pH 7.9), excised and stored at −80°C until DNA isolation.
DNA was isolated from lung and liver using a Wako DNA TIS Extractor kit (WAKO Chemicals USA, Richmond, VA, USA), according to the manufacturer’s recommendations with minor additions: 0.1mM desferoxamine mesylate (DFO) was added to the lysis solution, enzyme reaction solution and TE buffer, while 0.3mM DFO was added to the sodium hydroxide solution. DFO is an iron-chelating agent that prevents Fenton reactions and production of hydroxyl radicals that could artifactually oxidise DNA during sample preparation (20). Concentration and purity of DNA were determined by spectrophotometry.
Fifteen micrograms of DNA was digested per sample, using nuclease P1 and alkaline phosphatase (21). Briefly, 10 µl of 0.5M sodium acetate (pH 5.1) and 1 µl of 1M MgCl2 were added to 100 µl of 0.15mg/ml DNA. To denature genomic DNA, samples were heated at 100°C for 5min, then cooled on ice for 5min. One µl of nuclease P1 enzyme (Sigma N8630) was then added and the DNA samples were incubated for 1h at 37°C. Solution pH was adjusted to 7.8 by adding 1.25 µl of 1M Tris (pH 10.5). Ten µl of 0.1U/µl porcine alkaline phosphatase were added to each sample. Samples were then further incubated for 1h at 37°C. Enzymes were precipitated by adding 2 µl of 5.8M acetic acid, then samples were transferred to 10000 NMWL filter units and centrifuged (12000 × g for 20min at 4°C) to separate digested nucleosides from enzymes.
The concentration of oxidised guanine in digested liver and lung DNA was determined by interpolation using an eight-point standard curve (R 2 = 0.997±0.0015) generated using a commercially available DNA Oxidative Damage Enzyme-linked Immuno-Assay (EIA) kit (Cayman Chemical, Ann Arbor, MI). The antibody supplied with the DNA oxidative Damage EIA kit does not differentiate between 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), 8-oxoguanosine (8-oxoGuo) and 8-oxoguanine (8-oxoGua), so the assay product is referred to as oxidised guanine. However, the hydrolysis step in the DNA digestion process should only yield 2′-nucleosides, and the 8-oxoGuo should be removed by the RNase step during DNA digestion, leaving just 8-oxodG. The results presented have been corrected for weight of DNA digested.
Lung tumour induction and diagnosis
Fifty female mice per genotype were treated with either 50mg/kg AFB1 or with 40 µl DMSO ip. Mice that developed skin ulcerations or prolonged seizures, as well as those that lost > 15% of pre-study body weights, were humanely killed prior to study end. All remaining mice were killed after 75 weeks. Mice were killed by CO2 inhalation and suspect individual lung and liver masses ≥ 1mm in diameter were counted, measured and excised with the aid of a dissecting microscope. A portion of each mass was frozen in liquid nitrogen and stored at −80°C, and the remainder was fixed in 10% neutral buffered formalin. The liver and the remainder of the lungs were also excised; a portion of each liver and the left lobe of each lung were fixed in 10% neutral buffered formalin, and the remainder of the organs were frozen in liquid nitrogen prior to storage at −80°C. Evaluation of formalin-fixed, paraffin-embedded sections stained with hematoxylin and eosin was carried out by a pathologist (PVT) without knowledge of animal treatment and genotype.
K-ras mutation analysis
DNA was isolated from frozen lung tumours with a Qiagen DNeasy Blood and Tissue Kit (Qiagen, Toronto ON), according to the manufacturer’s recommendations. Exon 1 of K-ras, which contains codons 12 and 13, and Exon 2 of K-ras, which includes codon 61, were amplified by PCR (14).
Size of PCR products was confirmed by electrophoresis in 3% agarose gels. The remaining PCR products were then purified using a PCR Purification Kit (Qiagen, Toronto, ON, Canada), according to the manufacturer’s instructions. The purified PCR products were sequenced by the TCAG Sequencing Facility (Toronto, ON, Canada) using the forward PCR primers for K-ras exon 1 and exon 2.
Data analysis
Results are expressed as mean ± SD. To assess differences in oxidatively damaged DNA, a two-way analysis of variance (ANOVA) followed by a Bonferonni post hoc test was used. To assess weight changes, an unpaired t-test was used when comparing two groups and a one-way ANOVA followed by a Student Newman–Keuls post hoc was employed when comparing three or more groups. Tumour multiplicity was analysed using a Mann–Whitney U-test and genotype effects were compared using a two-way ANOVA with a Bonferonni post hoc test. For analysis of survival and mass incidence with respect to time, a Log-Rank (Mantel–Cox) test was used. Differences in the numbers of mice with lung or liver tumours and the frequencies of K-ras mutations in lung tumours between DMSO and AFB1-treated mice were analysed using Fisher’s exact test. In all cases, analysis was performed using Prism software (version 5.00) with statistical significance defined as P < 0.05.
Results
Oxidatively damaged DNA
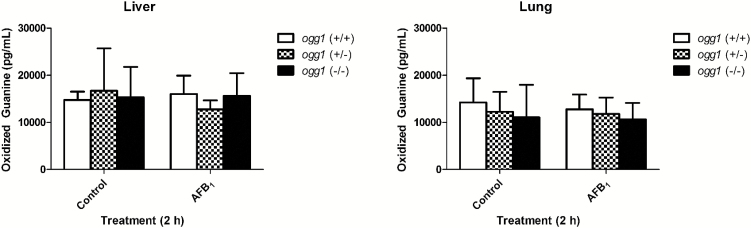
Levels of oxidised guanine 2h post-DMSO treatment did not differ between ogg1 genotypes in lung or liver, (P > 0.05, Figure 1). In addition, 50mg/kg of AFB1 did not increase the concentration of oxidatively damaged DNA compared with vehicle control 2h post-treatment, regardless of ogg1 genotype or tissue (P > 0.05, Figure 1).
Figure 1.
Concentration of oxidised guanine in control and AFB1-treated mouse liver and lung in ogg1 wild type, heterozygous and null mice. No significant differences were observed in oxidised guanine concentrations across ogg1 genotypes or due to AFB1 treatment, P > 0.05 (two-way ANOVA followed by Bonferonni post hoc test, n = 4).
Health status of mice in tumour study
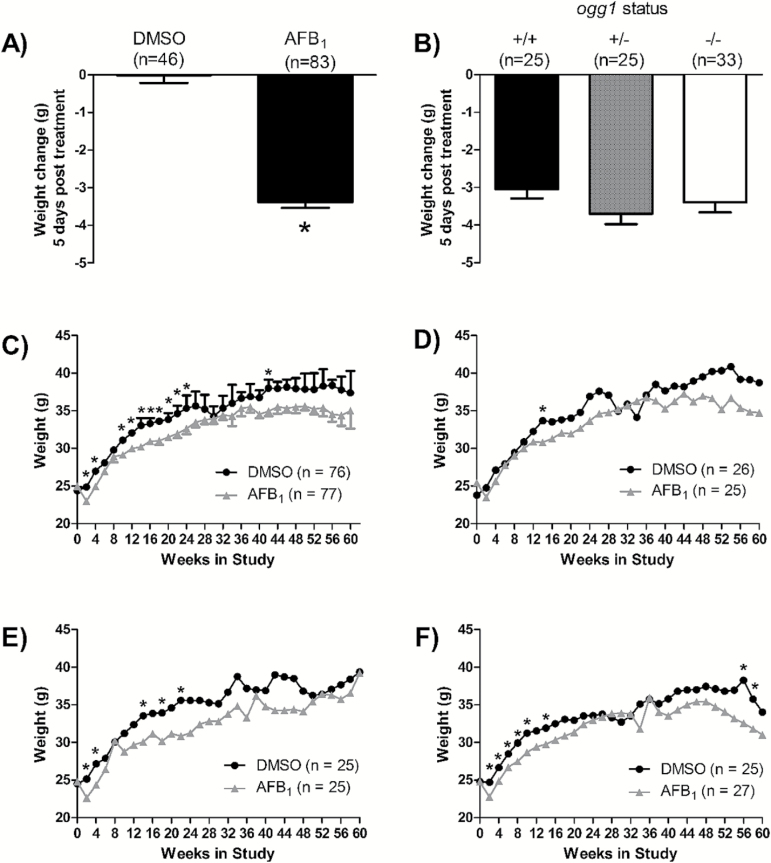
All mice treated with AFB1 had significant weight loss 5 days post-treatment, whereas weights of control animals were unaffected (Figure 2A). In addition, 9 of 36 ogg1 (−/−) mice treated with AFB1 were removed from the study within the first week post-treatment due to signs of dehydration and sluggishness. This only occurred in AFB1-treated ogg1 (−/−) mice. As such, ogg1 (−/−) mice treated with AFB1 were given sterile saline subcutaneously on days 3, 5, 7, 9 and 11 post-treatment. Once this fluid regimen was started, all ogg1 (−/−) mice survived treatment. The amount of AFB1-induced weight loss did not differ between ogg1 genotypes (Figure 2B); however, ogg1 (−/−) mice needed sterile saline to survive, suggesting that without the intervention the ogg1 (−/−) mice would have lost significant weight and potentially have died.
Figure 2.
Mouse weights throughout the tumourigenesis study. (A) weight change 5 days post treatment in all mice treated with DMSO or AFB1 compared with time 0; (B) weight change 5 days post-treatment in AFB1-treated ogg1 (+/+), (+/−) and (−/−) mice, compared with time 0; (C) comparison of weights of all control and all treated mice, regardless of genotype; weights of control and treated: (D) ogg1 (+/+) mice; (E) ogg1 (+/−) mice; and (F) ogg1 (−/−) mice. Error bars have been removed from graphs D–F for clarity. Significantly different from DMSO control at that time point (*P < 0.05, unpaired t-test (A, C–F); P > 0.05, one-way ANOVA with a Newman–Keuls post hoc test (B).
Following the initial weight loss 5 days post-treatment, both treated and control mice gained weight throughout the study. When the data for all genotypes were combined, AFB1-treated mice weighed on average ~2g less than control mice for the first 24 weeks, but recovered to similar weights for the remaining 51 weeks (Figure 2C). The number of time points that treated mice weighed less than control mice varied between genotypes. AFB1-treated ogg1 (+/+) mice weighed less than the ogg1 (+/+) controls at one time point, AFB1-treated ogg1 (+/−) mice weighed less than controls of the same genotype at five time points, and the AFB1-treated ogg1 (−/−) mice weighed significantly less than the ogg1 (−/−) control mice at eight time points, indicating a genotype related treatment effect for weight (Figure 2D–F). No difference in weight was observed between the three ogg1 genotypes in control mice (P > 0.05), or the three ogg1 genotypes in AFB1-treated mice (P > 0.05), emphasising that genotype alone did not affect mouse weight (data not shown).
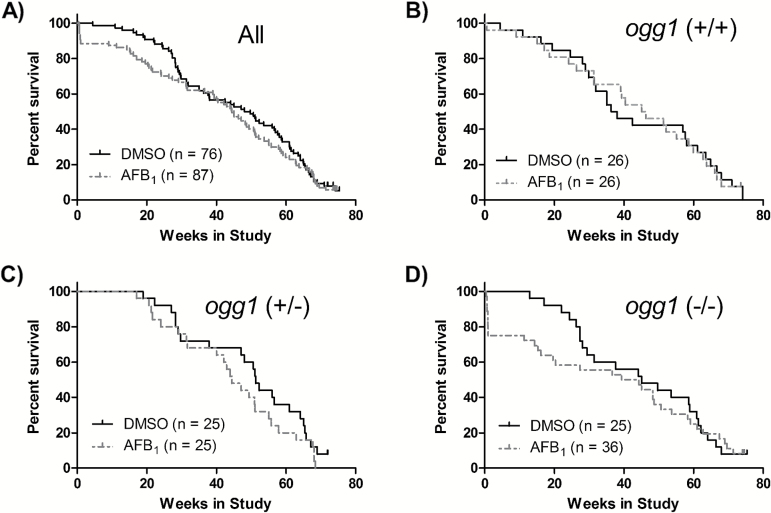
Over the course of the entire study, survival was not different between control and AFB1-treated mice, regardless of ogg1 genotype (Figure 3). ogg1 genotype also did not affect survival of control mice (P > 0.05, data not shown).
Figure 3.
Overall survival, expressed as the percentage of mice surviving in a week in: (A) all mice; (B) ogg1 (+/+) mice; (C) ogg1 (+/−) mice and (D) ogg1 (−/−) mice. No significant differences in survival were observed (P > 0.05, Mantel–Cox test).
Tumour induction
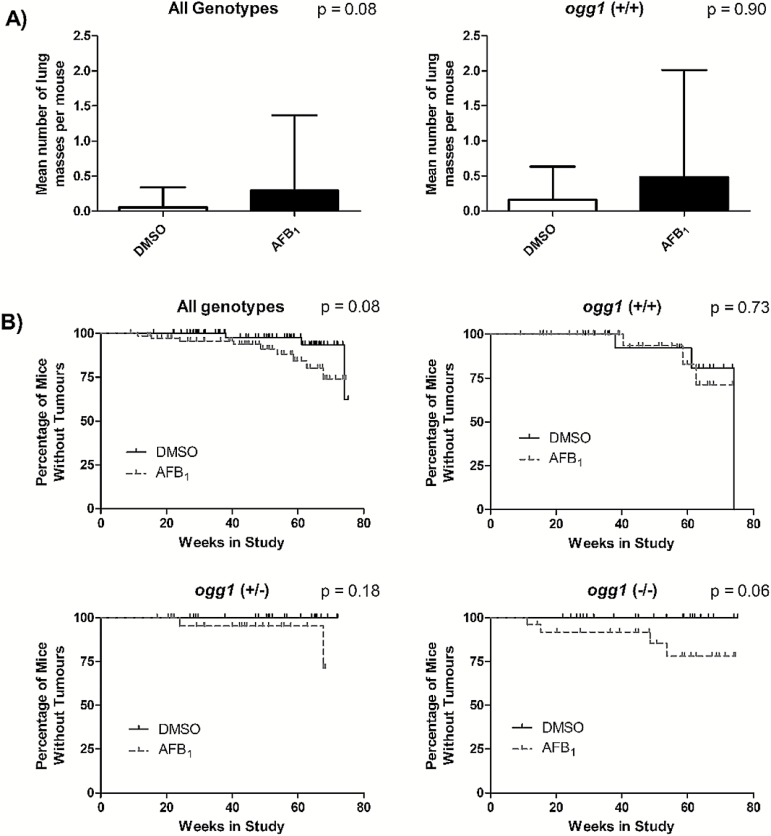
At the completion of the study, a total of 30 lung masses and 10 liver masses ≥ 1mm in diameter were excised. More lung and liver masses appeared to occur in AFB1-treated animals than in DMSO controls, but the differences were not significant (Table 1). Mean lung mass multiplicity appeared to be greater in the AFB1-treated mice versus controls, (0.29±1.08 lung masses/mouse and 0.06±0.29 lung masses/mouse respectively), but this was not significant (P = 0.08; Figure 4A). Lung mass multiplicity also was not different between control and treated when comparing just ogg1 (+/+) mice (0.16±0.47 lung masses/mouse and 0.48±1.5 lung masses/mouse, respectively; Figure 4A). Low numbers of lung masses precluded analysis within ogg1 (+/−) and ogg1 (−/−) mice.
Table 1.
Summary of the number of lung and liver masses and tumours in control and AFB1-treated micea
| Treatment | Ogg1 genotype | No. of mice with lung massesb | Total no. of lung massesc | No. of mice with lung tumoursd | Total no. of lung tumoursc | No. of mice with liver masses | Total no. of liver massesc | No. of mice with liver tumours | Total no. of liver tumoursc |
|---|---|---|---|---|---|---|---|---|---|
| DMSO | Wild type | 3/25 (12) | 4/25 | 1/25 (4) | 1/25 | 0/25 (0) | 0/25 | 0/25 (0) | 0/25 |
| Heterozygous | 0/25 (0) | 0/25 | 0/25 (0) | 0/25 | 1/25 (4) | 1/25 | 0/25 (0) | 0/25 | |
| Null | 0/25 (0) | 0/25 | 0/25 (0) | 0/25 | 0/25 (0) | 0/25 | 0/25 (0) | 0/25 | |
| All | 3/75 (4) | 4/75 | 1/75 (1) | 1/75 | 1/75 (1) | 1/75 | 0/75 (0) | 0/75 | |
| AFB1 e | Wild type | 3/25 (12) | 12/25 | 2/25 (8) | 10/25 | 1/25 (4) | 3/25 | 1/25 (4) | 3/25 |
| Heterozygous | 2/25 (8) | 6/25 | 1/25 (4) | 5/25 | 2/25 (8) | 4/25 | 2/25 (8) | 4/25 | |
| Null | 4/27 (19) | 4/27 | 2/27 (7) | 2/27 | 1/27 (4) | 2/27 | 1/27 (4) | 2/27 | |
| All | 9/77 (12) | 22/77 | 5/77 (6) | 17/77 | 4/77 (5) | 9/77 | 4/77 (5) | 9/77 |
AFB1, aflatoxin B1; DMSO, dimethyl sulfoxide.
aNumbers in parentheses are percentages.
bMasses refers to foci of hyperplasia, inflammation, adenoma, adenocarcinoma or hepatocellular carcinoma.
cNumbers represent the total number of lung or liver masses or tumours per group of mice.
dTumours refers to adenoma, adenocarcinoma or hepatocellular carcinoma.
eNo difference in the number of mice with lung or liver masses or tumours between AFB1 and DMSO, Fisher’s exact test, P > 0.05.
Figure 4.
Lung mass incidence and multiplicity. (A) Lung mass multiplicity following in vivo treatment with AFB1 in all genotypes combined and ogg1 (+/+) mice (P > 0.05, Mann–Whitney U-test). (B) Lung mass incidence with respect to time in all ogg1 genotypes combined, ogg1 (+/+) mice, ogg1 (+/−) mice and ogg1 (−/−) mice (P > 0.05, Mantel–Cox test).
Overall, a trend towards AFB1-treated mice developing lung masses earlier than control mice was apparent, but fell short of significance (P = 0.08; Figure 4B). No significant differences were observed between AFB1 treated and control mice for each genotype individually (Figure 4B). Of all of the control mice, only control ogg1 (+/+) mice developed lung masses (Table 1). When comparing ogg1 genotypes within AFB1-treated mice only, no difference in time of lung mass development was observed (data not shown).
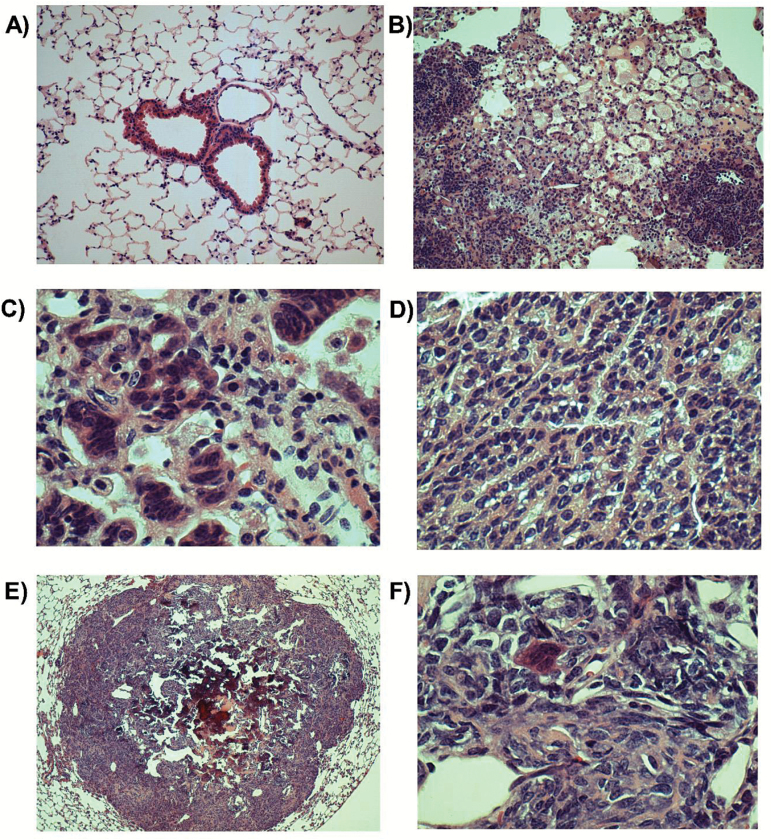
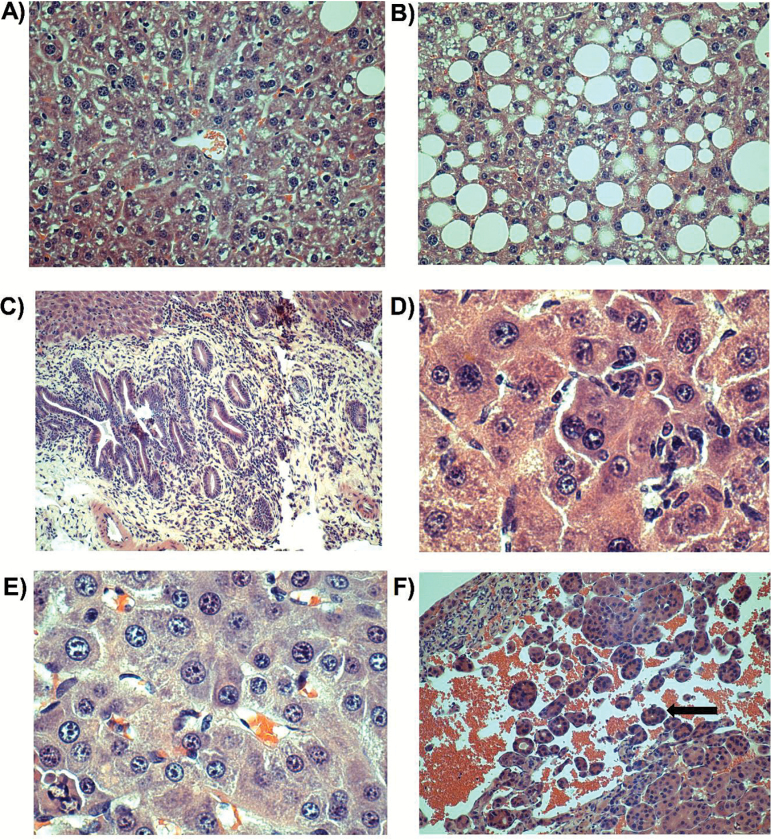
At the completion of the study, suspect lung masses from 12 mice were evaluated microscopically (three DMSO-treated mice and nine AFB1-treated mice). Of the suspect lung masses from DMSO-treated mice, those from two of the mice consisted of foci of alveolar hyperplasia or inflammation, and that from the other mouse was a pulmonary adenoma. Of the suspect lung masses from AFB1-treated mice, those from four of the mice were inflammatory foci, those from three of the mice were adenomas, and those from two of the mice were osteosarcomas (from an ogg1 (+/+) mouse and from an ogg1 (+/−) mouse). No osteosarcomas were observed in lung masses from DMSO-treated mice. Representative photomicrographs are shown in Figure 5. Five liver tumours were also evaluated microscopically (one from a DMSO-treated mouse and four from AFB1-treated mice). The liver from the DMSO-treated mouse had localised angiectasis surrounded by hepatocytes undergoing marked fatty degeneration. The liver masses from AFB1-treated mice were uniformly complex and often mixed tumours were present. The four liver masses from AFB1-treated mice had hepatocellular adenomas, and one also had a hepatocellular carcinoma (from ogg1 (+/+) mouse). Other microscopic findings in livers from these animals included altered cellular foci, scattered hepatocyte atypia and bile ductule proliferation. Representative photomicrographs are shown in Figure 6.
Figure 5.
Representative photomicrographs of lungs with corresponding magnification and treatment: (A) normal (×100; AFB1); (B) inflammation (×100; AFB1); (C) hyperplasia (×400; DMSO); (D) adenoma (×400; DMSO); (E) osteosarcoma (×40; AFB1); (F) osteosarcoma (×400; AFB1).
Figure 6.
Representative photomicrographs of livers with corresponding magnification and treatment: (A) normal (×200; DMSO); (B) generalised fatty degeneration of hepatocytes (×20; DMSO); (C) bile duct proliferation (×100; AFB1); (D) scattered hepatocyte atypia (×400; AFB1); (E) adenoma (×400; AFB1) and (F) hepatocellular carcinoma (×100; AFB1). Arrow is pointing towards an acinar pattern (little nest of malignant cells).
K-ras mutation analysis in lung tumours
Of the 30 lung masses excised, 18 were large enough for K-ras mutation analysis. K-ras mutations were observed in 3 of the 4 masses from DMSO-treated mice and in 3 of the 14 masses from AFB1-treated mice (Table 2). There was no statistical difference between the number of masses with K-ras mutations observed in control versus treated mice.
Table 2.
Summary of K-ras mutations in AFB1-induced and spontaneous OGG1 mouse lung massesa
| Treatment | ogg1 genotype | No. of masses with K-ras mutationsb | Codon 12 (normal = GGT) | Codon 61 (normal = CAA)c | ||
|---|---|---|---|---|---|---|
| GAT | CGA | CTA | CAC | |||
| DMSO | Wild type | 3/3 | 1 | 1 | 1 | 0 |
| Heterozygous | 0/1 | 0 | 0 | 0 | 0 | |
| Null | 0/0 | 0 | 0 | 0 | 0 | |
| All | 3/4 | 1 | 1 | 1 | 0 | |
| AFB1 d | Wild type | 2/8 | 0 | 0 | 0 | 2e |
| Heterozygous | 0/3 | 0 | 0 | 0 | 0 | |
| Null | 1/3 | 0 | 1e | 0 | 0 | |
| All | 3/14 | 0 | 1 | 0 | 2 | |
AFB1, aflatoxin B1; DMSO, dimethyl sulfoxide.
aNo mutations were found in K-ras codon 13.
bOf masses analysed for K-ras mutations (not all masses were available to analyse).
cToo few masses to compare mutation pattern statistically in specific K-ras codons.
dNo difference in number of K-ras mutations between AFB1 and DMSO, Fisher’s exact test, P > 0.05.
ePotential mutation, signal was faint.
Discussion
Previous studies have indicated that AFB1 can induce oxidatively damaged DNA, specifically 8-oxodG lesions, both in animal models and in humans (11,22). Additionally, acute exposure to a tumourigenic dose of AFB1 causes an increase in BER in the lung, which is correlated with an increase in OGG1 protein levels (11). Since adult mice develop lung tumours following exposure to AFB1, oxidatively damaged DNA, specifically 8-oxodG lesions, may be a contributing factor in AFB1-induced tumourigenesis. To test this hypothesis, we employed a mouse model deficient in OGG1, the rate-limiting enzyme in the removal of 8-oxodG lesions.
The observed lack of effect of ogg1 genotype on levels of 8-oxodG in lungs and livers of control mice is consistent with an earlier report (23). However, oxidatively damaged DNA, as measured by sensitivity to formamidopyrimidine DNA glycosylase, a bacterial enzyme that can excise 8-oxoGua lesions, was increased in ogg1 knockout mouse lung compared with wild-type mice, and the amount of oxidatively damaged DNA increased with age (23,24). Therefore, a difference in oxidatively damaged DNA may have been observed using a different method of detection, or differences in levels of oxidised guanine between ogg1 genotypes perhaps would have been observed with older mice. AFB1 treatment also did not affect the levels of oxidised guanine in lung or liver tissues 2-h post-treatment. These results contrast with previous studies that have shown that 8-oxodG levels increased 2-h post-treatment with the same dose of AFB1 (10,11). It is possible that the increase in 8-oxodG in this study occurred at an earlier or later time point than the 2-h time point measured, occurred only within specific lung cell types, or occurred in tissues other than the lung or liver. It is also possible that the hybrid mice employed are less sensitive to production of 8-oxodG lesions than mouse strains previously utilised.
Human exposure to AFB1 occurs most commonly through ingestion of AFB1-contaminated foods. As such, human exposure to AFB1 is often to low doses over a long period of time, which differs from the acute exposure protocol used in this study. However, the AFB1 treatment regimen employed in the current study is a tumourigenic dose that produces AFB1-DNA adducts that occur following human exposure to AFB1 (1,25). By employing this model of AFB1 toxicity, the results of this study can be directly compared with the results of previous studies investigating the mechanisms of AFB1 toxicity that employed the same dosing regimen.
Based on previous tumourigenesis studies employing AFB1, a single ip dose of 50mg/kg AFB1 to adult mice causes exclusively lung tumours, with up to 100% incidence in susceptible strains (13,14). The initial weight loss we observed in all mice after AFB1 treatment, regardless of genotype, and the mortality in the ogg1 (−/−) mice that occurred within one week of AFB1 treatment, were unexpected results. However, previous research indicates that efficiency of food use is consistently less in animals exposed to AFB1 than those not exposed (26). In addition, aflatoxin exposure was related to stunting and low body weight in children in Benin and Togo (26). The specificity of early AFB1-induced mortality for ogg1 (−/−) mice (Figure 3) suggests that OGG1 plays an important role in protecting tissues from damage induced by AFB1 exposure. Since OGG1 repairs 8-oxodG lesions, it is likely that increased oxidatively damaged DNA was at least partly responsible for the increased mortality in ogg1 (−/−) mice. As ogg1 (−/−) mice showed signs of dehydration, and administration of sterile saline to the ogg1 (−/−) mice alleviated the toxicity associated with AFB1 exposure, perhaps the gastrointestinal tract had increased oxidatively damaged DNA and water absorption was impaired. An alternative possibility is that the ogg1 (−/−) mice drank less water than ogg1 (+/−) or ogg1 (+/+) mice, a behaviour that is controlled by the hypothalamus. Due to the unanticipated and late-occurring dehydration, coupled with the necessity to hydrate the animals parenterally, assessment of fluid intake was not possible. However, OGG1 is expressed and active in the hypothalamus of rats (27), allowing for the possibility that oxidatively damaged DNA was significantly increased in the hypothalamus of AFB1-treated ogg1 (−/−) mice, which could cause alterations in the expression of genes that regulate water consumption. A final possibility could be perturbed fluid regulation due to AFB1-induced oxidatively damaged DNA in the kidney.
After the first week following treatment, overall survival was not affected by either AFB1 treatment or ogg1 genotype, and although initially AFB1-treated mice weighed less than DMSO-treated mice, this effect was lost as the study progressed, indicating a transient toxicity of AFB1. It was interesting, however, that ogg1 (−/−) and ogg1 (+/−) mice weighed less than wild-type controls at more time points than did the AFB1-treated ogg1 (+/+) mice. This result suggests that without both OGG1 alleles, the mice were slower to recover from damage caused by AFB1.
Overall, tumour incidence was much lower in this study than in previous AFB1-induced tumourigenesis studies (13,14). One possibility for this outcome is the mouse model employed, given that C57Bl/6 mice are relatively resistant and sv129 mice are of intermediate susceptibility to lung tumourigenesis. When considering the genotypes individually, no significant effects were observed between treated and control mice, which may have been a result of the low numbers of lung and liver masses.
Adult mice are resistant to AFB1-induced hepatocarcinogenicity, a phenomenon attributable at least in part to constitutive expression of glutathione S-transferases in mouse liver, which have high catalytic activity toward detoxifying the AFB1-epoxide (28–31). In addition, mouse liver nucleotide excision repair activity is about 6-fold higher than rat liver nucleotide excision repair activity, which also correlates with species susceptibility to AFB1-induced liver tumours (32). Since liver tumours were only observed following AFB1 treatment, and occurred at similar frequencies for all three ogg1 genotypes, this result was not attributable to deletion of ogg1. In the few liver tumours that were excised from AFB1-treated mice, many interesting pathologies were found. Notable was the presence of hepatocellular carcinoma, which is the human lesion commonly associated with chronic exposure to AFB1 (25,33).
It was surprising that two of the AFB1-treated mice had multiple osteosarcomas in their lungs. Indeed, certain DNA repair syndromes in humans are associated with an increased incidence of osteosarcomas, (34) and we have demonstrated previously that AFB1 perturbs DNA repair in mice (32). However, the low osteosarcoma incidence (2 of 75 AFB1-treated mice) precludes drawing conclusions regarding cause-and-effect.
Results of the K-ras mutation analysis were consistent with the lung masses in DMSO-treated mice being spontaneous, as anticipated. In addition, lung masses from AFB1-treated mice did not have the characteristic G to T transversion mutations in codon 12 of K-ras that are associated with AFB1-induced mouse lung tumours (13,35).
Inactivating mutations in the OGG1 gene have been documented in a small number of sporadic human lung, kidney and gastric tumours (36,37). In this study, ogg1 genotype did not appear to have a significant effect on AFB1-induced tumourigenesis, although due to low numbers of lung and liver masses, a definitive conclusion cannot be made. The apparent lack of effect could be due to the possibility that 8-oxodG lesions are not major contributors to AFB1-induced tumourigenesis. Another possibility is that a genotype-specific effect to the susceptibility of AFB1 tumourigenesis was not observed due to the particular hybrid mice employed, as mouse strains vary in their susceptibility to AFB1-induced effects. Finally, the lack of effect could potentially indicate that even without OGG1, adaptive processes were able to compensate for any increase in 8-oxodG lesions. For example, in the absence of OGG1, translesion DNA synthesis can contribute to protecting the cell from 8-oxodG residues (38).
In conclusion, mice with compromised ability to repair oxidised DNA did not have increased sensitivity to AFB1-induced oxidatively damaged DNA and tumourigenesis, suggesting that oxidatively damaged DNA may not be a major contributing process in AFB1-induced tumourigenicity. However, deletion of one or both alleles of ogg1 did increase susceptibility to other toxic effects of AFB1.
Funding
This work was supported by the Canadian Institutes of Health Research (MOP-89798).
Acknowledgements
We thank Ms Sandra Graham for technical assistance.
Conflict of interest statement: None declared.
References
- 1. Dvorackova I., Stora C., Ayraud N. (1981). Evidence of aflatoxin B1 in two cases of lung cancer in man. J. Cancer Res. Clin. Oncol., 100, 221–224. [DOI] [PubMed] [Google Scholar]
- 2. Harrison J. C., Garner R. C. (1991). Immunological and HPLC detection of aflatoxin adducts in human tissues after an acute poisoning incident in S.E. Asia. Carcinogenesis, 12, 741–743. [DOI] [PubMed] [Google Scholar]
- 3. Hayes R. B., van Nieuwenhuize J. P., Raatgever J. W., ten Kate F. J. (1984). Aflatoxin exposures in the industrial setting: an epidemiological study of mortality. Food Chem. Toxicol., 22, 39–43. [DOI] [PubMed] [Google Scholar]
- 4. Garner R. C., Miller E. C., Miller J. A. (1972). Liver microsomal metabolism of aflatoxin B 1 to a reactive derivative toxic to Salmonella typhimurium TA 1530. Cancer Res., 32, 2058–2066. [PubMed] [Google Scholar]
- 5. Hertzog P. J., Smith J. R., Garner R. C. (1982). Characterisation of the imidazole ring-opened forms of trans-8,9-dihydro-8,9-dihydro-8-(7-guanyl)9-hydroxy aflatoxin B1. Carcinogenesis, 3, 723–725. [DOI] [PubMed] [Google Scholar]
- 6. Nakabeppu Y., Sakumi K., Sakamoto K., Tsuchimoto D., Tsuzuki T., Nakatsu Y. (2006). Mutagenesis and carcinogenesis caused by the oxidation of nucleic acids. Biol. Chem., 387, 373–379. [DOI] [PubMed] [Google Scholar]
- 7. Valko M., Rhodes C. J., Moncol J., Izakovic M., Mazur M. (2006). Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact., 160, 1–40. [DOI] [PubMed] [Google Scholar]
- 8. David S. S., O’Shea V. L., Kundu S. (2007). Base-excision repair of oxidative DNA damage. Nature, 447, 941–950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Boiteux S., Radicella J. P. (2000). The human OGG1 gene: structure, functions, and its implication in the process of carcinogenesis. Arch. Biochem. Biophys., 377, 1–8. [DOI] [PubMed] [Google Scholar]
- 10. Guindon K. A., Bedard L. L., Massey T. E. (2007). Elevation of 8-hydroxydeoxyguanosine in DNA from isolated mouse lung cells following in vivo treatment with aflatoxin B(1). Toxicol. Sci., 98, 57–62. [DOI] [PubMed] [Google Scholar]
- 11. Guindon-Kezis K. A., Mulder J. E., Massey T. E. (2014). In vivo treatment with aflatoxin B1 increases DNA oxidation, base excision repair activity and 8-oxoguanine DNA glycosylase 1 levels in mouse lung. Toxicology, 321, 21–26. [DOI] [PubMed] [Google Scholar]
- 12. Wang D., Kreutzer D. A., Essigmann J. M. (1998). Mutagenicity and repair of oxidative DNA damage: insights from studies using defined lesions. Mutat. Res., 400, 99–115. [DOI] [PubMed] [Google Scholar]
- 13. Guindon K. A., Foley J. F., Maronpot R. R., Massey T. E. (2008). Failure of catalase to protect against aflatoxin B1-induced mouse lung tumorigenicity. Toxicol. Appl. Pharmacol., 227, 179–183. [DOI] [PubMed] [Google Scholar]
- 14. Donnelly P. J., Devereux T. R., Foley J. F., Maronpot R. R., Anderson M. W., Massey T. E. (1996). Activation of K-ras in aflatoxin B1-induced lung tumors from AC3F1 (A/J x C3H/HeJ) mice. Carcinogenesis, 17, 1735–1740. [DOI] [PubMed] [Google Scholar]
- 15. Christmann M., Tomicic M. T., Roos W. P., Kaina B. (2003). Mechanisms of human DNA repair: an update. Toxicology, 193, 3–34. [DOI] [PubMed] [Google Scholar]
- 16. Slupphaug G., Kavli B., Krokan H. E. (2003). The interacting pathways for prevention and repair of oxidative DNA damage. Mutat. Res., 531, 231–251. [DOI] [PubMed] [Google Scholar]
- 17. Hazra T. K., Das A., Das S., Choudhury S., Kow Y. W., Roy R. (2007). Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair (Amst)., 6, 470–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Klungland A., Rosewell I., Hollenbach S., Larsen E., Daly G., Epe B., Seeberg E., Lindahl T., Barnes D. E. (1999). Accumulation of premutagenic DNA lesions in mice defective in removal of oxidative base damage. Proc. Natl. Acad. Sci. USA, 96, 13300–13305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wong A. W., McCallum G. P., Jeng W., Wells P. G. (2008). Oxoguanine glycosylase 1 protects against methamphetamine-enhanced fetal brain oxidative DNA damage and neurodevelopmental deficits. J. Neurosci., 28, 9047–9054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ravanat J. L. (2005). Measuring oxidized DNA lesions as biomarkers of oxidative stress: an analytical challenge. FABAD J. Pharm. Sci., 30, 100–113. [Google Scholar]
- 21. Gupta N., Curtis R. M., Mulder J. E., Massey T. E. (2013). Acute in vivo treatment with 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone does not alter base excision repair activities in murine lung and liver. DNA Repair (Amst)., 12, 1031–1036. [DOI] [PubMed] [Google Scholar]
- 22. Peng T., Li L. Q., Peng M. H., et al. (2007). Evaluation of oxidative stress in a group of adolescents exposed to a high level of aflatoxin B1–a multi-center and multi-biomarker study. Carcinogenesis, 28, 2347–2354. [DOI] [PubMed] [Google Scholar]
- 23. Risom L., Møller P., Dybdahl M., Vogel U., Wallin H., Loft S. (2007). Dietary exposure to diesel exhaust particles and oxidatively damaged DNA in young oxoguanine DNA glycosylase 1 deficient mice. Toxicol. Lett., 175, 16–23. [DOI] [PubMed] [Google Scholar]
- 24. Osterod M., Hollenbach S., Hengstler J. G., Barnes D. E., Lindahl T., Epe B. (2001). Age-related and tissue-specific accumulation of oxidative DNA base damage in 7,8-dihydro-8-oxoguanine-DNA glycosylase (Ogg1) deficient mice. Carcinogenesis, 22, 1459–1463. [DOI] [PubMed] [Google Scholar]
- 25. Eaton D. L., Gallagher E. P. (1994). Mechanisms of aflatoxin carcinogenesis. Annu. Rev. Pharmacol. Toxicol., 34, 135–172. [DOI] [PubMed] [Google Scholar]
- 26. Williams J. H., Phillips T. D., Jolly P. E., Stiles J. K., Jolly C. M., Aggarwal D. (2004). Human aflatoxicosis in developing countries: a review of toxicology, exposure, potential health consequences, and interventions. Am. J. Clin. Nutr., 80, 1106–1122. [DOI] [PubMed] [Google Scholar]
- 27. Verjat T., Dhénaut A., Radicella J. P., Araneda S. (2000). Detection of 8-oxoG DNA glycosylase activity and OGG1 transcripts in the rat CNS. Mutat. Res., 460, 127–138. [DOI] [PubMed] [Google Scholar]
- 28. Buetler T. M., Eaton D. L. (1992). Complementary DNA cloning, messenger RNA expression, and induction of alpha-class glutathione S-transferases in mouse tissues. Cancer Res., 52, 314–318. [PubMed] [Google Scholar]
- 29. Hayes J. D., Judah D. J., Neal G. E., Nguyen T. (1992). Molecular cloning and heterologous expression of a cDNA encoding a mouse glutathione S-transferase Yc subunit possessing high catalytic activity for aflatoxin B1-8,9-epoxide. Biochem. J., 285, 173–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. McDonagh P. D., Judah D. J., Hayes J. D., Lian L. Y., Neal G. E., Wolf C. R., Roberts G. C. (1999). Determinants of specificity for aflatoxin B1-8,9-epoxide in alpha-class glutathione S-transferases. Biochem. J., 339, 95–101. [PMC free article] [PubMed] [Google Scholar]
- 31. Wong Z. A., Hsieh D. P. (1980). The comparative metabolism and toxicokinetics of aflatoxin B1 in the monkey, rat, and mouse. Toxicol. Appl. Pharmacol., 55, 115–125. [DOI] [PubMed] [Google Scholar]
- 32. Bedard L. L., Alessi M., Davey S., Massey T. E. (2005). Susceptibility to aflatoxin B1-induced carcinogenesis correlates with tissue-specific differences in DNA repair activity in mouse and in rat. Cancer Res., 65, 1265–1270. [DOI] [PubMed] [Google Scholar]
- 33. Busby W. F., Wogan G. N. (1984). Aflatoxin. In Searle CE. (ed.), Chemical Carcinogens, 2 edn American Chemical Society, Washington, DC, pp. 945–1136. [Google Scholar]
- 34. Thoms K. M., Kuschal C., Emmert S. (2007). Lessons learned from DNA repair defective syndromes. Exp. Dermatol., 16, 532–544. [DOI] [PubMed] [Google Scholar]
- 35. Donnelly P. J., Massey T. E. (1999). Ki-ras activation in lung cells isolated from AC3F1 (A/J x C3H/HeJ) mice after treatment with aflatoxin B1. Mol. Carcinog., 26, 62–67. [DOI] [PubMed] [Google Scholar]
- 36. Chevillard S., Radicella J. P., Levalois C., Lebeau J., Poupon M. F., Oudard S., Dutrillaux B., Boiteux S. (1998). Mutations in OGG1, a gene involved in the repair of oxidative DNA damage, are found in human lung and kidney tumours. Oncogene, 16, 3083–3086. [DOI] [PubMed] [Google Scholar]
- 37. Shinmura K., Kohno T., Kasai H., Koda K., Sugimura H., Yokota J. (1998). Infrequent mutations of the hOGG1 gene, that is involved in the excision of 8-hydroxyguanine in damaged DNA, in human gastric cancer. Jpn. J. Cancer Res., 89, 825–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yasui M., Kanemaru Y., Kamoshita N., Suzuki T., Arakawa T., Honma M. (2014). Tracing the fates of site-specifically introduced DNA adducts in the human genome. DNA Repair (Amst)., 15, 11–20. [DOI] [PubMed] [Google Scholar]