Figure 2.
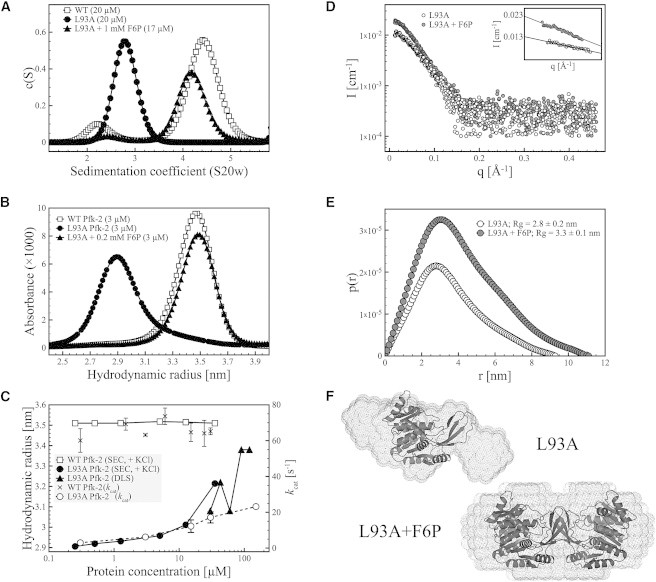
Hydrodynamic properties of the L93A mutant of Pfk-2. (A) Sedimentation velocity experiments show that the L93A mutant (black circles) is a monomer at low protein concentration. Addition of F6P leads to substrate-induced dimerization of the L93A mutant (black triangles) with a sedimentation coefficient close to the native dimer of Pfk-2 (open squares). (B) Analytical SEC of L93A Pfk-2 showing that the L93A mutant is a monomer (black circles) with a hydrodynamic radius of 2.9 nm, which can be guided toward formation of a dimer with a hydrodynamic radius of 3.5 nm upon addition of F6P (black triangles). This value is close to the hydrodynamic radius estimated for the native dimer (open squares). (C) Dependence of the hydrodynamic properties and enzyme activity of L93A Pfk-2 on the protein concentration, measured by SEC and DLS. The hydrodynamic radius of the L93A mutant (black circles and triangles) becomes larger upon increasing protein concentrations, suggesting a shift in the monomer-dimer equilibrium, whereas the value of 3.5 nm estimated for the native dimer (white squares) remains constant at all protein concentrations assayed. The increase in hydrodynamic radius of L93A is accompanied by an increase in enzyme activity (open circles), having a kcat that is about threefold lower than the WT enzyme (black crosses) at the highest protein concentration assayed (150 μM). (D) SAXS data of L93A in the absence (open circles) and presence (gray circles) of F6P. The inset shows the linear fitting of q values <1.3/Rg according to the Guinier approximation. The difference in I0 between both samples is about twofold, consistent with a change in the oligomeric state of L93A. (E) Interatomic distance distribution function p(r) of L93A in the absence (open circles) and presence (gray circles) of F6P. The estimated radius of gyration of the substrate-induced dimer of L93A in the presence of F6P is larger than the monomer, in agreement with SEC and sedimentation velocity experiments. (F) Ab initio shape models of L93A with and without F6P superimposed to the crystal structure of the native dimer and the compact monomer of Pfk-2 (PDB ID 3N1C), respectively.
