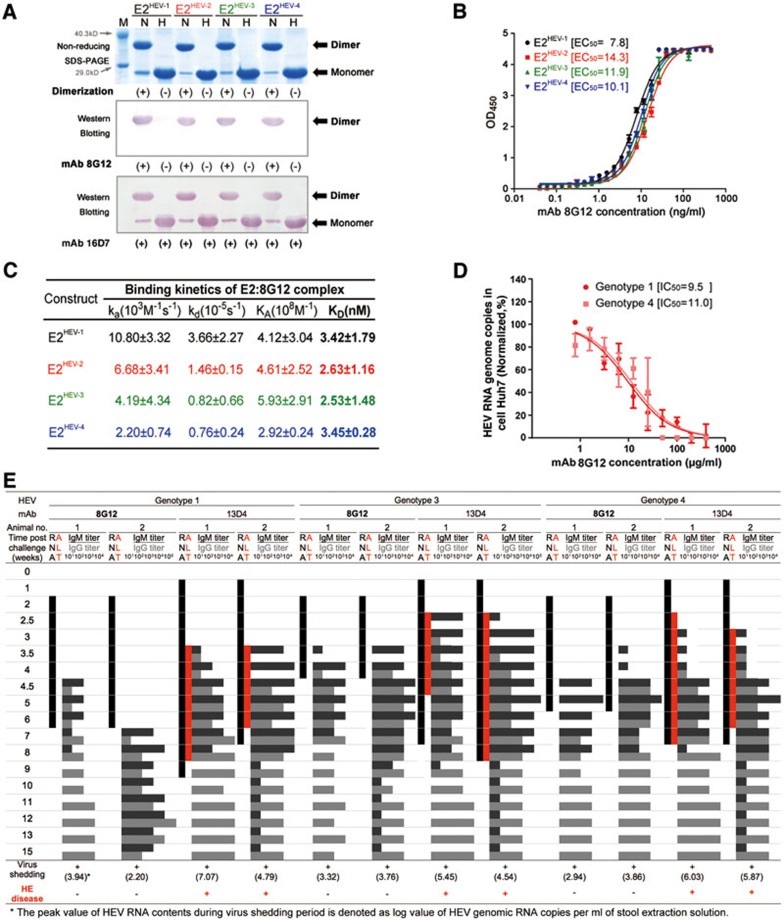
Figure 1.
Comparable reactivity and neutralization nature of mAb 8G12 is observed among various HEV genotypes. (A) E2 proteins with four genotypes were subjected to non-reducing SDS-PAGE and western blot analysis with the neutralizing mAb 8G12 to investigate their reactivity in association with each genotype. The lanes marked with H indicate heated samples under reducing conditions (samples were heated up to 100 °C for 3 min) whereas the lanes marked with N indicate samples under non-reducing conditions (0.1% SDS, in the absence of β-mercaptoethanol (BME) and not heated). (+) denotes dimerization or reactivity with 8G12, (−) denotes monomer or loss of the respective property. (B) The interaction of E2 proteins with four genotypes against mAb 8G12 was tested by ELISA and EC50 was calculated by sigmoid trend fitting. (C) The affinity constants of the interaction of E2 proteins with four genotypes against mAb 8G12 were measured by SPR in Biacore 3000. (D) Inhibition profiles of mAb 8G12 blocking authentic HEV from infecting the host cell. HEV genotypes 1 or 4 show 1 × 105 genomic RNA copies in infected vulnerable Huh7 cells in the presence of serial dilutions of mAb 8G12. The inhibition profiles of HEV RNA genome copies vs. concentration of mAb 8G12 were fitted to a sigmoid trend to generate the IC50 value. (E) Neutralization of HEV infection rate by mAb 8G12 in a Macaques rhesus monkey model. HEV of genotypes 1, 3 and 4 was mixed with mAb 8G12 or mAb 13D4 (negative control) and used to inoculate monkeys. Serum and stool samples were taken before and weekly after infection to monitor serum levels of alanine aminotransferase (ALT; red bar), shedding of the HEV RNA in stool samples (black bar) and HEV IgG/IgM antibody titer (gray histogram). (+) denotes occurrence of virus shedding or Hepatitis E disease, (−) denotes a negative event. Also refer to Supplementary information, Figure S5.