Abstract
Background
KRAS mutation occurs in 35%-40% of colorectal cancer (CRC). The aim of our study was to evaluate the pathological and molecular features of specific KRAS mutated colorectal carcinomas. KRAS and BRAFV600E mutation tests were performed in 762 primary tumors from a consecutive cohort study of Chinese CRC patients.
Methods
DNA mismatch repair (MMR) status was determined by immunohistochemistry (IHC) staining. Assessment of KRAS and BRAF V600E mutational status was performed using a multiplex allele-specific PCR-based assay.
Results
Mutations of KRAS (34.8%) and BRAFV600E (3.1%) were nearly mutually exclusive. Both KRAS- and BRAF- mutated tumors were more likely to be located at proximal colon than wild-type (WT) carcinomas. KRAS-mutated carcinomas were more frequently observed in female patients (47.5% vs 37.1%, p = 0.005) and mucinous differentiation (34.7% vs 24.8%, p = 0.004), but have no difference between lymph node (LN) metastases and among pTNM stages. Whereas, BRAF-mutated carcinomas more frequently demonstrated histologic features such as proximal location (60.9% vs 20.9%, p = 0.001), low-grade histology (43.5% vs 18.0%, p = 0.005), mucinous differentiation (69.6% vs 25.9%, p = 0.001) and deficient MMR (dMMR) (21.7% vs 7.6%, p = 0.03). In particular, KRAS codon 12 mutated carcinomas had increased lymph node metastasis (odds ratio [OR] = 1.31; 95% confidence interval [CI] = 1.04 to 1.65; P = 0.02) and were more likely in higher disease stage (III-IV) than that of WT carcinomas (OR = 1.30; 95% CI = 1.03 to 1.64; P = 0.03). However, there were no significant differences in lymph node metastasis and disease stage between KRAS codon 13 mutated carcinoma and WT carcinoma patients.
Conclusions
In summary, KRAS codon 12 mutation, but not codon 13 mutation, is associated with lymph node metastasis and higher tumor stages.
Keywords: KRAS mutation, BRAF mutation, Colorectal cancer, Codon 12 and 13, DNA mismatch repair
Background
Colorectal cancer (CRC) is the third most common cancer and the fourth leading cause of cancer mortality in China [1]. CRC is a multistep process based on the accumulation of somatic mutations and can be divided into at least two different and seemingly independent pathways, which is the chromosomal instability (CIN) and microsatellite instability (MSI) pathways [2-5]. CIN occurs in about 85% patients with sporadic CRC and is thought to originate from a relatively uniform and linear accumulation of genetic changes in APC, KRAS and TP53 genes [6]. However, sporadic tumors with MSI-high (MSI-H) are originated from promoter hypermethylation of the MLH1 gene, more frequently found in females, and tend to be poorly differentiated tumors, of mucinous subtype and often harboring somatic mutations in BRAFV600E [7,8]. Dysfunction of the DNA mismatch repair (MMR) system is the main cause of MSI, which leads to accelerated accumulation of single nucleotide mutations and alterations in the length of simple, repetitive microsatellite sequences [9].
Recently, MMR status, KRAS and BRAF mutation status have attracted remarkable attention due to their potential prognostic and predictive role in colorectal carcinomas [10-12]. KRAS mutations are present in approximately 35% to 40% of colon cancers, with roughly 2/3 of these mutations in codon 12 and 1/3 in codon 13 [11,13]. The presence of a KRAS mutation is predictive for resistance to anti-EFGR monoclonal antibodies (mAbs) in advanced colon cancer [13-16]. However, the biological and functional consequences of KRAS mutations at codon 12 may be different from those at codon 13 [17-19]. It has been suggested that patients whose tumors harbor a KRAS Gly13Asp mutation may benefit from anti-EGFR mAb therapy [20-22]. The clinical significance of KRAS mutation in colorectal carcinoma patients is controversial; some studies reported no association with survival, whereas others suggested that patients with KRAS mutated colorectal carcinoma have poorer outcome for any mutation subtype, mutation in codon 12 only or codon 13 only [19,21,23,24].
On the other hand, BRAF is also involved in the MAPK/ERK signaling pathway and oncogenic mutations in this gene have been identified in CRC [25]. Several studies have reported a range of frequencies regarding BRAF mutations in colon cancer (7.1%–13.3%), with the most frequent mutation being a single substitution at nucleotide 1,799, substituting valine for glutamic acid (V600E) [7]. Mutations in BRAF are most commonly found in microsatellite-unstable (MSI) tumors, whereas they are less common in microsatellite-stable (MSS) tumors [10,26,27]. Mutations in KRAS and BRAF genes seem to occur in a mutually exclusive manner, and both are suggested as integral components for an effective molecular classification of colorectal cancer.
More accurate prediction of outcome among patients with CRC remains a worthy area of investigation. Although the roles of MMR status, KRAS and BRAF mutations on clinical outcome are frequently documented, the accurate analysis of these 3 features on clinicopathologic and prognosis with emphasis on the specific KRAS gene mutation is still missing. The aim of this study was to evaluate the prognostic role of MMR status, BRAF mutations and specific KRAS point mutation in 762 patients in Chinese population, and several clinicopathologic features to better stratify colorectal cancer patients.
Methods
Study population
The clinicopathological records of 762 patients with corresponding paraffin-embedded material available for molecular analysis were retrospectively collected from the Department of Pathology, Cancer Institute and Hospital, Chinese Academy of Medical Sciences, Beijing, China from December 2011 to December 2012. Patients who had a history of preoperative radiochemotherapy or gastrointestinal surgical resection were excluded. Histopathological criteria were reviewed and included tumor diameter, pT and pN classification, grade of differentiation, histological subtype, tumor location, tumor size as well as the pTNM stage. The size of each tumor was evaluated by measuring its maximum diameter. Grading was determined according to the 2010 WHO histological classification. The pTNM stage system of the 7th edition AJCC cancer staging was used. Evaluating of M stage was mainly according to confirmed pathological results and/or radiological data. Location in the colon was designated as proximal colon for tumors located in the cecum, ascending colon and transverse colon, and as distal colon for tumors in the descending colon and sigmoid colon. Mucinous differentiation in the tumor was defined by the presence of pools of extracellular mucin-containing clusters of carcinomatous cells. When >50% of analyzed tumor demonstrated mucinous differentiation, the tumor was classified as mucinous carcinoma. The study was approved by the Institute Review Board of the Cancer Hospital, CICAMS. Each participant signed an Institutional Review Board approved informed consent in accordance with current guidelines.
KRAS and BRAFV600E mutation analysis
Assessment of KRAS and BRAF V600E mutational status was performed in the Molecular Pathology Laboratory of Department of Pathology, CICAMS, using appropriate quality control procedures. Mutation status was determined using genomic DNA extracted from macrodissected formalin-fixed, paraffin-embedded tumor tissue. Both KRAS (codons 12 and 13) and BRAF (p.V600E) mutation tests were performed using a multiplex allele-specific PCR-based assay (ACCB, Beijing, China), together with the Stratagene Mx3000P (Agilent Technologies Inc, Santa Clara, CA), which assesses seven different potential mutations in KRAS codons 12 and 13 (Gly12Ala, Gly12Asp, Gly12Arg, Gly12Cys, Gly12Ser, Gly12Val, and Gly13Asp). Neither KRAS nor BRAFV600E mutated tumors were designated as WT carcinomas.
DNA mismatch repair proteins expression
A panel of four-antibody of MMR proteins was performed as a routine practice in our pathological department, contained MLH1, PMS2, MSH2 and MSH6. All of the 762 samples were stained in an autostainer (Autostainer Link 48, Dako, Denmark). Primary mouse monoclonal antibodies included MLH1 antibody (ES05, Dako), MSH2 antibody (FE11, Dako). Primary rabbit monoclonal antibodies included MSH6 antibody (EP49, Dako) and PMS2 antibody (EP51, Dako). Carcinomas were considered as deficient MMR (dMMR) when there was a completely absent staining of a detectable nuclear signal in neoplastic cells for at least one protein. While the adjacent normal mucosa or stromal/lymphoid cells that showed presence of nuclear staining are regarded as internal positive control.
Statistical analysis
The primary objective of this study was to identify distinct clinicopathologic features associated with specific KRAS and BRAFV600E mutation status. Differences of patient characteristics and clinicopathologic factors in the two-dimensional cross-comparison were evaluated statistically by Pearson’s χ2-test or Fischer’s exact test. Statistical tests were two-sided, and P < 0.05 were considered significant. Logistic regression models were used to detect associations of these characteristics with each of the specific KRAS mutations and provided estimates of odds ratio (ORs) and confidence intervals (CIs). Statistics were carried out using SPSS software (version 16.0 of SPSS, Chicago, IL, USA).
Results
Primary samples from 762 colorectal carcinoma patients were analyzed for KRAS, BRAF gene mutations and MMR status. Mutations of KRAS occurred in 34.8% of colorectal carcinomas. BRAFV600E mutation was demonstrated in 3.1% of colorectal carcinomas. There was one tumor demonstrating mutations in both KRAS and BRAF; this case was excluded from the analysis. Mutated carcinomas were compared with non mutated carcinomas for sex, age, histological features and molecular characteristics (Table 1). In addition, given that non-KRAS-mutated tumors include a distinct subset characterized by BRAF mutation, analyses were also performed to compare KRAS-mutated tumors with both BRAF-mutated tumors and the remaining subset of colorectal carcinomas, with observed neither somatic oncogene mutation.
Table 1.
Distributions of clinicopathologic characteristics by KRAS and BRAF mutation status
| Characterics | MutantKRAS | Wild-typeKRAS | P-value | MutantBRAF | Wild-typeBRAF | P-value |
|---|---|---|---|---|---|---|
| (n = 265) | (n = 496) | (n = 23) | (n = 738) | |||
| Sex | 0.005 | 0.26 | ||||
| Male | 139 (52.5%) | 312 (62.9%) | 11 (47.8%) | 440 (59.6%) | ||
| Female | 126 (47.5%) | 184 (37.1%) | 12 (52.2%) | 298 (40.4%) | ||
| Tumor location | 0.004 | <0.0001 | ||||
| Proximal colon | 73 (28.3%) | 90 (18.7%) | 14 (60.9%) | 149 (20.9%) | ||
| Distal colon | 52 (20.2%) | 135 (28.0%) | 5 (21.7%) | 178 (25.0%) | ||
| Rectum | 133 (51.5%) | 257 (53.3%) | 4 (17.4%) | 386 (54.1%) | ||
| pT stage | 0.39 | 0.35‡ | ||||
| pT1-2 | 31 (11.7%) | 69 (13.9%) | 1 (4.3%) | 98 (13.3%) | ||
| pT3-4 | 234 (88.3%) | 427 (86.1%) | 22 (95.7%) | 640 (86.7%) | ||
| pN stage | 0.09 | 0.11 | ||||
| pN0 | 124 (46.8%) | 264 (53.2%) | 8 (34.8%) | 380 (51.5%) | ||
| pN1-2 | 141 (53.2%) | 232 (46.8%) | 15 (65.2%) | 358 (48.5%) | ||
| Disease stage | 0.10 | 0.13 | ||||
| I-II | 123 (46.2%) | 260 (52.4%) | 8 (34.8%) | 374 (50.7%) | ||
| III-IV | 143 (53.8%) | 236 (47.6%) | 15 (65.2%) | 364 (49.3%) | ||
| Tumor grade | 0.26 | 0.005‡ | ||||
| G1-2 | 221 (83.4%) | 397 (80.0%) | 13 (56.5%) | 605 (82.0%) | ||
| G3 | 44 (16.6%) | 99 (20.0%) | 10 (43.5%) | 133 (18.0%) | ||
| Histological type | 0.004 | <0.0001‡ | ||||
| Mucinous | 92 (34.7%) | 123 (24.8%) | 16 (69.6%) | 191 (25.9%) | ||
| Ohter | 173 (65.3%) | 373 (75.2%) | 7 (30.4%) | 547 (74.1%) | ||
| MMR status | 0.76 | 0.03§ | ||||
| Proficient | 243 (91.7%) | 458 (92.3%) | 18 (78.3%) | 682 (92.4%) | ||
| Deficient | 22 (8.3%) | 38 (7.7%) | 5 (21.7%) | 56 (7.6%) | ||
| Age, y | 0.65† | 0.35† | ||||
| Mean (SD) | 57.7 ± 11.3 | 57.3 ± 11.5 | 59.6 ± 10.1 | 57.3 ± 11.4 | ||
| Median | 58.5 | 58.0 | 59.0 | 58.0 | ||
| Range | 27.0-84.0 | 21.0-87.0 | 38.0-82.0 | 21.0-87.0 | ||
| Age,y | 0.95 | 0.23‡ | ||||
| <45 | 37 (14.0%) | 70 (14.1%) | 1 (4.3%) | 105 (14.2%) | ||
| ≥45 | 228 (86.0%) | 426 (85.9%) | 22 (95.7%) | 633 (85.8%) | ||
| Tumor size (cm) | 0.68† | 0.004† | ||||
| Mean (SD) | 4.95 ± 2.08 | 4.89 ± 1.81 | 5.83 ± 2.13 | 4.87 ± 1.89 | ||
| Median | 4.5 | 4.5 | 6 | 4.5 | ||
| Range | 1.5-17.0 | 1.0-14.0 | 3.0-11.0 | 1.0-17.0 | ||
| Tumor size | 0.32 | 0.09 | ||||
| <4.5 cm | 121 (45.7%) | 208 (41.9%) | 6 (26.1%) | 322 (43.6%) | ||
| ≥4.5 cm | 144 (54.3%) | 288 (58.1%) | 17 (73.9%) | 416 (56.4%) |
Abbreviations: MMR mismatch repair, SD standard deviation.
†Two-sided Kruskal Wallis test.
‡Two-sided χ2 test with continuity correction.
§Fischer’s exact test.
Others are two-sided χ2 test.
Clinical information and morphological characteristics
The mean age at presentation for KRAS-mutated carcinoma was 57.7 ± 11.3 years, which was no significantly different to that for non-mutated-KRAS carcinoma at 57.3 ± 11.5 years and WT carcinoma cases at 57.1 ± 11.5 years. Furthermore, regarding the age, there was no significant difference in younger (<45) and older (≥45) patients among KRAS-mutated, KRAS-wild type and BRAF-mutated carcinomas.
Gender distribution did not differ significantly between BRAF-mutated colorectal carcinomas. However, female patients were more likely to possess KRAS mutation than males (47.7% vs 37.1%, P = 0.004). BRAF-mutated carcinomas were more likely to be found in proximal colon than wild-type BRAF tumors (60.9% vs 20.9%, P < 0.0001). In addition, compared with KRAS-wild type carcinomas, KRAS-mutated carcinomas were more likely located in proximal colon (28.3% vs 18.7%, P = 0.004). Twenty five subjects had synchronous carcinomas in both proximal colon and rectum, including 6 KRAS-mutated and 19 KRAS-wild type carcinoma. However, there was no synchronous carcinoma found in BRAF-mutated patients in our study. In addition, BRAF-mutated carcinomas were significantly associated with larger tumor size compared with wild-type BRAF tumors (5.83 ± 2.13 vs 4.87 ± 1.89; P = 0.004).
Both BRAF- and KRAS-mutated carcinomas demonstrated more frequently mucinous differentiation when compared with BRAF- and KRAS- wild type carcinomas respectively (34.7% vs 24.8% and 69.6% vs 25.9%; P = 0.004 for KRAS-mutated carcinomas vs KRAS-wild type carcinomas; P < 0.0001 for BRAF-mutated carcinomas vs BRAF-wild type carcinomas). BRAF-mutated carcinomas were observed with higher tumor grade (G3) than BRAF-wild type carcinomas (43.5% vs 18.0%; P = 0.005), however, this did not differ significantly with KRAS-mutated and -wild type carcinomas. In addition, there were no significantly difference in aspects of pT stage, pN stage and disease stage among KRAS-mutated, BRAF-mutated and wild type carcinomas.
Differences with specific KRAS mutations in CRC
The distribution and frequencies of the various tumor and clinicopathological characteristics of specific KRAS mutation were summarized in Table 2. Mutation frequencies at codon 12 and codon 13 were 26.9% (205/761) and 7.9% (60/761), respectively. The most common variant in codon 12 was the p.G12D/A mutation (101/265, 38.1%), followed by the p.G12V mutation (64/265, 24.2%). The p.G13/D mutation frequency was 22.6% (60/265). There was no gender preponderance between codon 13 mutation and WT carcinoma patients. However, female patients were more likely to carry codon 12 mutations than WT carcinoma cases (47.8% vs 36.4%; OR = 1.38; 95% CI = 1.10 to 1.74; P = 0.005). Codon 12 and 13 mutated carcinomas were more likely to be found in the proximal location than WT carcinomas respectively (Codon 12: 29.1% vs 16.7%; OR = 3.17; 95% CI = 2.24 to 4.49; P < 0.0001. Codon 13: 25.0% vs 16.7%; OR = 3.37; 95% CI = 1.67 to 6.82; P < 0.0001). Both codon 12 and codon 13 mutated carcinomas demonstrated more frequently mucinous differentiation when compared with WT carcinomas (Codon 12: 33.7% vs 21.1%; OR = 1.53; 95% CI = 1.21 to 1.93; P = 0.001; Codon 13: 36.7% vs 21.1%; OR = 1.95; 95% CI = 1.20 to 3.17; P = 0.001).
Table 2.
Distributions of clinicopathologic characteristics by KRAS codon 12 and codon 13 mutation
| Characterics | MutantKRASCodon 12 | Null carcinoma | P-value (Codon 12vs Null) | MutantKRASCodon 13 | Pvalue (Codon 13vs Null) |
|---|---|---|---|---|---|
| (n = 205) | (n = 473) | (n = 60) | |||
| Sex | 0.005 | 0.12 | |||
| Male | 107 (52.2%) | 301 (63.6%) | 32 (53.3%) | ||
| Female | 98 (47.8%) | 172 (36.4%) | 28 (46.7%) | ||
| Tumor location | <0.0001 | <0.0001 | |||
| Proximal colon | 58 (29.1%) | 76 (16.7%) | 15 (25.0%) | ||
| Distal colon | 40 (20.1%) | 253 (55.7%) | 13 (21.7%) | ||
| Rectum | 101 (50.8%) | 125 (27.6%) | 32 (53.3%) | ||
| pT stage | 0.20 | 0.89 | |||
| pT1-2 | 22 (10.7%) | 68 (14.4%) | 9 (15.0%) | ||
| pT3-4 | 183 (89.3%) | 405 (85.6%) | 51 (85.0%) | ||
| pN stage | 0.02 | 0.90 | |||
| pN0 | 91 (44.4%) | 256 (54.1%) | 33 (55.0%) | ||
| pN1-2 | 114 (55.6%) | 217 (45.9%) | 27 (45.0%) | ||
| Disease stage | 0.03 | 0.99 | |||
| I-II | 90 (43.9%) | 252 (53.3%) | 32 (53.3%) | ||
| III-IV | 115 (56.1%) | 221 (46.7%) | 28 (46.7%) | ||
| Tumor grade | 0.14 | 0.26‡ | |||
| G1-2 | 176 (85.9%) | 384 (81.2%) | 45 (75.0%) | ||
| G3 | 29 (14.1%) | 89 (18.8%) | 15 (25.0%) | ||
| Histological type | 0.001 | 0.007 | |||
| Mucinous | 69 (33.7%) | 100 (21.1%) | 22 (36.7%) | ||
| Ohter | 136 (66.3%) | 373 (78.9%) | 38 (63.3%) | ||
| MMR status | 0.87 | 0.30‡ | |||
| Proficient | 190 (92.7%) | 440 (93.0%) | 53 (88.3%) | ||
| Deficient | 15 (7.3%) | 33 (7.0%) | 7 (11.7%) | ||
| Age, y | 0.59† | 0.47† | |||
| Mean (SD) | 57.6 ± 11.4 | 57.1 ± 11.5 | 58.3 ± 10.6 | ||
| Median | 59.0 | 58.0 | 57.5 | ||
| Range | 27.0-84.0 | 21.0-87.0 | 31.0-77.0 | ||
| Age,y | 0.86 | 0.09 | |||
| <45 | 31 (15.1%) | 69 (14.6%) | 4 (6.7%) | ||
| ≥45 | 174 (84.9%) | 404 (85.4%) | 56 (93.3%) | ||
| Tumor size (cm) | 0.27 | 0.96 | |||
| Mean (SD) | 4.99 ± 2.13 | 4.82 ± 1.76 | 4.83 ± 1.92 | ||
| Median | 4.5 | 4.5 | 4.4 | ||
| Range | 1.5-17.0 | 1.0-14.0 | 3.5-12.0 | ||
| Tumor size | 0.77 | 0.28 | |||
| <4.5 cm | 90 (43.9%) | 202 (42.7%) | 30 (50.0%) | ||
| ≥4.5 cm | 115 (56.1%) | 271 (57.3%) | 30 (50.0%) |
Abbreviations: MMR mismatch repair, SD standard deviation, Null neither KRAS nor BRAFV600E mutation.
†Two-sided Kruskal Wallis test.
‡Two-sided χ2 test with continuity correction.
Others are two-sided χ2 test.
Univariate logistic regression models identified the following factors as statistically significantly between KRAS codon 12 mutated carcinomas and WT carcinomas: gender, pN stage, pTNM stage and histological subtype (Figure 1A). In particular, codon 12 mutated carcinomas had increased lymph node metastasis (pN stage) than WT carcinomas (55.6% vs 45.9%; OR = 1.31; 95% CI = 1.04 to 1.65; P = 0.02). Moreover, codon 12 mutated carcinomas were more likely in higher disease stages (III-IV) than that of WT carcinomas (56.1% vs 46.7%; OR = 1.30; 95% CI = 1.03 to 1.64; P = 0.02; P = 0.03). However, there were no significant differences in lymph node metastasis and disease stage between codon 13 mutated carcinomas and WT carcinoma patients (Figure 1B). In addition, KRAS mutated carcinomas demonstrated no differences in tumor invasion depth (pT stage) with WT carcinomas in both codon 12 and 13 mutated cases.
Figure 1.
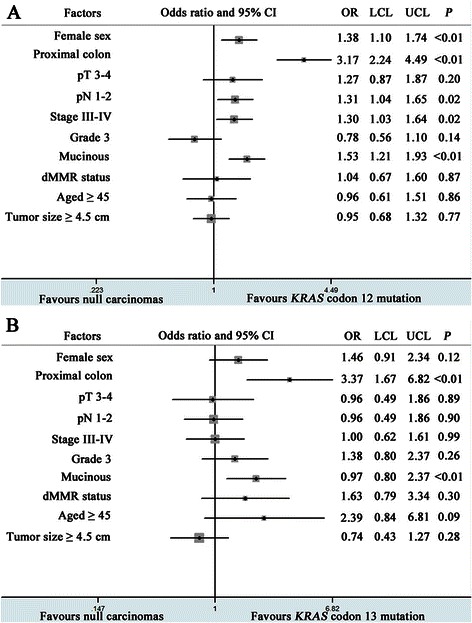
Forest plot of univariate logistic model associations with KRAS codon 12 mutation status (A) and codon 13 mutation status (B). P values are for two-sided Pearson χ2 test. CI = confidence interval; dMMR = deficient mismatch repair; LCL = lower confidence limit; UCL = upper confidence limit; OR = odds ratio.
Discussion
To the best of our knowledge, this is the first study to demonstrate that patients with KRAS codon 12 mutated colorectal carcinomas had a more advanced tumor stage than those with tumors harboring p.G13D mutation or wild-type KRAS in a large cohort of Chinese population. In addition, the KRAS codon 12 mutated carcinomas in our study also displayed morphologic features typically associated with adverse behavior regardless of tumor location. Our data are consistent with previous reports, indicating that the presence of a mutation in KRAS codon 12 confers substantially greater oncogenic potential than codon 13 mutation [18,20,28-30].
The incidence of KRAS mutation in our reports was approximately 34.8%, which is similar to that described in other studies. However, the frequency of BRAFV600E mutation was 3.1%, which is lower than in studies of western countries, but similar to that of Asian populations [27,31-33]. This indicates that ethnicity and/or environmental factors might contribute to the discrepancy. Unlike reports from Caucasians, patients with BRAFV600E mutated tumors in our study did not differ significantly in aspects of sex and age [10]. KRAS mutated cases were more likely to be female patients but with no significant difference in age distribution. Several studies reported that BRAFV600E mutated tumors were more likely to have a more aggressive biology with four or more positive lymph nodes and higher pTNM stage in large population-based cohort studies [34-37]. However, this observation was inconsistent with our results, which presented that mutation in either BRAFV600E or KRAS was not related to the adverse clinicopathological features accompanied by advanced lymph node metastasis and III-IV disease stages. Admittedly, this difference may be related to our smaller size of the BRAFV600E mutation subgroup. However, both BRAFV600E and KRAS mutated carcinomas were more likely to be located in the proximal colon and mucinous carcinomas. In addition, MMR deficient status was less likely to be associated with KRAS mutations, but have a positive correlation with BRAFV600E mutations [10,35]. Goldstein J, et al. reported that BRAFV600E mutation is a poor prognostic factor in metastatic MSI-H colorectal tumors [38].
To our knowledge based on the literature search in Pubmed, this is the first study to address the clinicopathological difference between KRAS codon 12 and 13 mutations in over 500 of BRAF-wild type colorectal cancers in Chinese population [39-41]. Although several previous studies had distinguished the difference between the prognostic associations of KRAS mutations in codon 12 and 13, none of the large studies (with a sample size more than 300) controlled for BRAF mutation status in their analyses, and results were conflicting [12,24,42,43]. The initial studies considered both mutations including codon 12 and 13 as a whole to analyze the clinicopathological features and disease outcome. Only small and very recent detailed reports assessed the effect of KRAS mutations where the codon 12 and 13 were analyzed separately [18,19,21]. RASCAL study, the initial data from clinical trials, suggested that KRAS mutation status is an important prognostic factor for progression and outcome in the CRC patients and glycine to valine in codon 12 convey a more aggressive biological manner [22,43,44]. The results have been broadly accepted all over the world except that the patients enrolled in the study were distributed over 21 different countries and the results may be confounded with the different ethnic and environmental factors.
In our study, compared with WT carcinoma cases, both KRAS codon 12 and codon 13 mutations were associated with proximal (vs distal) tumor site of the colon. The distribution of KRAS codon 12 and 13 mutations did not differ considerably by tumor subsite , consistent with findings from other reports [19]. Besides, proximal colon carcinomas were more likely than distal carcinomas to be KRAS-mutated and BRAF-mutated. Rosty. C, et al. reported that both KRAS- and BRAF-mutated carcinomas more frequently demonstrated focal or predominant mucinous differentiation than WT carcinomas, which was consistent with our findings [11]. In addition, mucinous tumors were more likely than other type of colorectal carcinomas in either KRAS codon 12 or 13 mutated tumors. Mucinous colonic adenocarcinoma is a frequently encountered histologic subtype of colorectal tumors, which is often associated with worse clinical outcome and decreased overall survival [45,46].
The most valuable finding of this study is that KRAS codon 12 mutated tumors demonstrated more positive lymph nodes and pTNM III-IV stage of disease than WT carcinoma patients, whereas tumors with codon 13 mutation did not differ by number of positive lymph nodes or pTNM stage. This was consistent with findings from a smaller report, which KRAS codon 12 mutation was found to be linked with more aggressive clinicopathological features and worse clinical outcomes [19,47]. Mutations in codons 12 and 13 lead to alterations in encoded amino acids adjacent to the GTP binding pocket and reduced the GTPase activity of KRAS protein after guanine nucleotide activating protein (GAP) binding [17,48-50]. Subsequently, these conformational and structural changes of the EGFR signaling pathway are out of control with constitutive activation of KRAS protein. Nonetheless, theses structural modifications will be different in case of each codon and the amino acid changed conferring variable activated KRAS effects. In particular, in vitro and in vivo studies indicate that KRAS codon 12 mutations have greater transforming capacity when compared with codon 13 mutations [23,51,52]. On the basis of protein computational analysis, codon 12-mutated KRAS remains in an active GTP-bound state longer than codon 13-mutated or wild type KRAS. It seems that mutations in codon 13 share the similar protein confirmation with wild type KRAS [53]. Consequently, codon 13 mutations confer reduced transforming ability in colon tumor cells.
Despite these positive findings, our study had some limitations. First, because our study lacked data on tumor CIMP (Cpg Island Methylator Phenotype) status and cause of dMMR status, we could not distinguish the correlations of somatic and germline mismatch repair mutations with KRAS and BRAF mutations. Second, although based on a large population size, the low incidences of BRAF mutation (3.1%) made the subgroups relatively small and further validations with more population are needed. Finally, we did not examine other less common mutations in KRAS codons 61, 117 and 146, which are also the negative predictive marker for response to anti-EGFR therapy [54].
Conclusions
In conclusion, our study suggests that specific epidemiologic and clinicopathologic characteristics were associated with KRAS and BRAFV600E mutations in a large cohort of Chinese colorectal carcinoma population. Specifically, tumor size of 6 cm or longer, low-grade histology, and dMMR status were associated with a higher incidence of BRAFV600E-mutated tumors. Both mutations tend to be proximal tumor location and mucinous histology, but KRAS-mutated tumors are more common in female patients. Finally, KRAS codon 12 mutation, but not codon 13 mutation, is associated with more positive lymph nodes and higher pTNM stages. Because of its correlation with a more advanced stage, patients with KRAS codon 12 mutations may have worse survival than those with KARS 13 mutations or wild-type KRAS. Further studies need to define the mechanism by the clinicopathologic and epidemiologic characteristics that may explain the association with these specific mutations.
Acknowledgements
We thank all study participants of the Department of Pathology for their contributions to this project. This work was supported by a grant from Youth Backbone Program (to Jianming Ying) of Cancer Hospital, CAMS, Beijing, China and National Nature Science Foundation of China (81401984).
Abbreviations
- CRC
Colorectal cancer
- MMR
Mismatch repair
- IHC
Immunohistochemistry
- OR
Odds ratio
- CIN
Chromosomal instability
- MSI
Microsatellite instability
- CIMP
Cpg island methylator phenotype
Footnotes
Wenbin Li and Tian Qiu contributed equally to this work.
Competing interests
The authors declare that they have no competing interests.
Authors’ contributions
JMY designed the study and drafted the manuscript. WBL analyzed data and drafted the manuscript. TQ carried out the molecular genetic studies and participated in the statistical analysis. YL and LS sequenced the alignment. WXZ, SSS and SMZ obtained patients’ pathological and clinical information. NL participated in the study design and the critical review of the manuscript. All authors read and approved the final manuscript.
Contributor Information
Wenbin Li, Email: liwenbin9631@hotmail.com.
Tian Qiu, Email: qiutian1228@126.com.
Wenxue Zhi, Email: zhiwenxue303@sina.com.
Susheng Shi, Email: shishusheng@sina.com.
Shuangmei Zou, Email: smzou@hotmail.com.
Yun Ling, Email: lingyun_1991@163.com.
Ling Shan, Email: shanling0612@hotmail.com.
Jianming Ying, Email: jmying@hotmail.com.
Ning Lu, Email: nlu03@126.com.
References
- 1.Siegel R, Desantis C, Jemal A. Colorectal cancer statistics, 2014. CA Cancer J Clin. 2014;64(2):104–17. doi: 10.3322/caac.21220. [DOI] [PubMed] [Google Scholar]
- 2.Cancer Genome Atlas Network Comprehensive molecular characterization of human colon and rectal cancer. Nature. 2012;487(7407):330–7. doi: 10.1038/nature11252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fearon ER. Molecular genetics of colorectal cancer. Annu Rev Pathol. 2011;6:479–507. doi: 10.1146/annurev-pathol-011110-130235. [DOI] [PubMed] [Google Scholar]
- 4.Ogino S, Chan AT, Fuchs CS, Giovannucci E. Molecular pathological epidemiology of colorectal neoplasia: an emerging transdisciplinary and interdisciplinary field. Gut. 2011;60(3):397–411. doi: 10.1136/gut.2010.217182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogino S, Goel A. Molecular classification and correlates in colorectal cancer. J Mol Diagn. 2008;10(1):13–27. doi: 10.2353/jmoldx.2008.070082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Frattini M, Balestra D, Suardi S, Oggionni M, Alberici P, Radice P, et al. Different genetic features associated with colon and rectal carcinogenesis. Clin Cancer Res. 2004;10(12 Pt 1):4015–21. doi: 10.1158/1078-0432.CCR-04-0031. [DOI] [PubMed] [Google Scholar]
- 7.Maestro ML, Vidaurreta M, Sanz-Casla MT, Rafael S, Veganzones S, Martinez A, et al. Role of the BRAF mutations in the microsatellite instability genetic pathway in sporadic colorectal cancer. Ann Surg Oncol. 2007;14(3):1229–36. doi: 10.1245/s10434-006-9111-z. [DOI] [PubMed] [Google Scholar]
- 8.Popat S, Hubner R, Houlston RS. Systematic review of microsatellite instability and colorectal cancer prognosis. J Clin Oncol. 2005;23(3):609–18. doi: 10.1200/JCO.2005.01.086. [DOI] [PubMed] [Google Scholar]
- 9.Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology. 2010;138(6):2073–87. doi: 10.1053/j.gastro.2009.12.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gonsalves WI, Mahoney MR, Sargent DJ, Nelson GD, Alberts SR, Sinicrope FA, et al. Patient and tumor characteristics and BRAF and KRAS mutations in colon cancer, NCCTG/Alliance N0147. J Natl Cancer Inst. 2014; 106(7). doi:10.1093/jnci/dju106. [DOI] [PMC free article] [PubMed]
- 11.Rosty C, Young JP, Walsh MD, Clendenning M, Walters RJ, Pearson S, et al. Colorectal carcinomas with KRAS mutation are associated with distinctive morphological and molecular features. Mod Pathol. 2013;26(6):825–34. doi: 10.1038/modpathol.2012.240. [DOI] [PubMed] [Google Scholar]
- 12.Zlobec I, Kovac M, Erzberger P, Molinari F, Bihl MP, Rufle A, et al. Combined analysis of specific KRAS mutation, BRAF and microsatellite instability identifies prognostic subgroups of sporadic and hereditary colorectal cancer. Int J Cancer. 2010;127(11):2569–75. doi: 10.1002/ijc.25265. [DOI] [PubMed] [Google Scholar]
- 13.Normanno N, Tejpar S, Morgillo F, De Luca A, Van Cutsem E, Ciardiello F. Implications for KRAS status and EGFR-targeted therapies in metastatic CRC. Nat Rev Clin Oncol. 2009;6(9):519–27. doi: 10.1038/nrclinonc.2009.111. [DOI] [PubMed] [Google Scholar]
- 14.Jonker DJ, O’Callaghan CJ, Karapetis CS, Zalcberg JR, Tu D, Au HJ, et al. Cetuximab for the treatment of colorectal cancer. N Engl J Med. 2007;357(20):2040–8. doi: 10.1056/NEJMoa071834. [DOI] [PubMed] [Google Scholar]
- 15.Karapetis CS, Khambata-Ford S, Jonker DJ, O’Callaghan CJ, Tu D, Tebbutt NC, et al. K-ras mutations and benefit from cetuximab in advanced colorectal cancer. N Engl J Med. 2008;359(17):1757–65. doi: 10.1056/NEJMoa0804385. [DOI] [PubMed] [Google Scholar]
- 16.Perkins G, Pilati C, Blons H, Laurent-Puig P. Beyond KRAS status and response to anti-EGFR therapy in metastatic colorectal cancer. Pharmacogenomics. 2014;15(7):1043–52. doi: 10.2217/pgs.14.66. [DOI] [PubMed] [Google Scholar]
- 17.Er TK, Chen CC, Bujanda L, Herreros-Villanueva M. Clinical relevance of KRAS mutations in codon 13: Where are we? Cancer Lett. 2014;343(1):1–5. doi: 10.1016/j.canlet.2013.09.012. [DOI] [PubMed] [Google Scholar]
- 18.Imamura Y, Morikawa T, Liao X, Lochhead P, Kuchiba A, Yamauchi M, et al. Specific mutations in KRAS codons 12 and 13, and patient prognosis in 1075 BRAF wild-type colorectal cancers. Clin Cancer Res. 2012;18(17):4753–63. doi: 10.1158/1078-0432.CCR-11-3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoon HH, Tougeron D, Shi Q, Alberts SR, Mahoney MR, Nelson GD, et al. KRAS codon 12 and 13 mutations in relation to disease-free survival in BRAF-wild-type stage III colon cancers from an adjuvant chemotherapy trial (N0147 alliance) Clin Cancer Res. 2014;20(11):3033–43. doi: 10.1158/1078-0432.CCR-13-3140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.De Roock W, Jonker DJ, Di Nicolantonio F, Sartore-Bianchi A, Tu D, Siena S, et al. Association of KRAS p.G13D mutation with outcome in patients with chemotherapy-refractory metastatic colorectal cancer treated with cetuximab. JAMA. 2010;304(16):1812–20. doi: 10.1001/jama.2010.1535. [DOI] [PubMed] [Google Scholar]
- 21.Mao C, Huang YF, Yang ZY, Zheng DY, Chen JZ, Tang JL. KRAS p.G13D mutation and codon 12 mutations are not created equal in predicting clinical outcomes of cetuximab in metastatic colorectal cancer: a systematic review and meta-analysis. Cancer. 2013;119(4):714–21. doi: 10.1002/cncr.27804. [DOI] [PubMed] [Google Scholar]
- 22.Russo A, Bazan V, Agnese V, Rodolico V, Gebbia N. Prognostic and predictive factors in colorectal cancer: Kirsten Ras in CRC (RASCAL) and TP53CRC collaborative studies. Ann Oncol. 2005;16(Suppl 4):iv44–9. doi: 10.1093/annonc/mdi907. [DOI] [PubMed] [Google Scholar]
- 23.Cerottini JP, Caplin S, Saraga E, Givel JC, Benhattar J. The type of K-ras mutation determines prognosis in colorectal cancer. Am J Surg. 1998;175(3):198–202. doi: 10.1016/S0002-9610(97)00283-3. [DOI] [PubMed] [Google Scholar]
- 24.Roth AD, Tejpar S, Delorenzi M, Yan P, Fiocca R, Klingbiel D, et al. Prognostic role of KRAS and BRAF in stage II and III resected colon cancer: results of the translational study on the PETACC-3, EORTC 40993, SAKK 60–00 trial. J Clin Oncol. 2010;28(3):466–74. doi: 10.1200/JCO.2009.23.3452. [DOI] [PubMed] [Google Scholar]
- 25.Fang JY, Richardson BC. The MAPK signalling pathways and colorectal cancer. Lancet Oncol. 2005;6(5):322–7. doi: 10.1016/S1470-2045(05)70168-6. [DOI] [PubMed] [Google Scholar]
- 26.French AJ, Sargent DJ, Burgart LJ, Foster NR, Kabat BF, Goldberg R, et al. Prognostic significance of defective mismatch repair and BRAF V600E in patients with colon cancer. Clin Cancer Res. 2008;14(11):3408–15. doi: 10.1158/1078-0432.CCR-07-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen D, Huang JF, Liu K, Zhang LQ, Yang Z, Chuai ZR, et al. BRAFV600E mutation and its association with clinicopathological features of colorectal cancer: a systematic review and meta-analysis. PLoS One. 2014;9(3) doi: 10.1371/journal.pone.0090607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lin CC, Lin JK, Lin TC, Chen WS, Yang SH, Wang HS, et al. The prognostic role of microsatellite instability, codon-specific KRAS, and BRAF mutations in colon cancer. J Surg Oncol. 2014;110(4):451–7. doi: 10.1002/jso.23675. [DOI] [PubMed] [Google Scholar]
- 29.Bazan V, Migliavacca M, Zanna I, Tubiolo C, Grassi N, Latteri MA, et al. Specific codon 13 K-ras mutations are predictive of clinical outcome in colorectal cancer patients, whereas codon 12 K-ras mutations are associated with mucinous histotype. Ann Oncol. 2002;13(9):1438–46. doi: 10.1093/annonc/mdf226. [DOI] [PubMed] [Google Scholar]
- 30.Winder T, Mundlein A, Rhomberg S, Dirschmid K, Hartmann BL, Knauer M, et al. Different types of K-Ras mutations are conversely associated with overall survival in patients with colorectal cancer. Oncol Rep. 2009;21(5):1283–7. doi: 10.3892/or_00000352. [DOI] [PubMed] [Google Scholar]
- 31.Tol J, Nagtegaal ID, Punt CJ. BRAF mutation in metastatic colorectal cancer. N Engl J Med. 2009;361(1):98–9. doi: 10.1056/NEJMc0904160. [DOI] [PubMed] [Google Scholar]
- 32.Qiu J, Compagnone M, Laibe S, Lagarde A, Goncalves A, Turrini O, et al. BRAF p.Val600Glu (V600E) somatic mutation is mainly associated with MSS phenotype in metastatic colorectal cancer. Cancer Genomics Proteomics. 2011;8(1):15–8. [PubMed] [Google Scholar]
- 33.Yuan ZX, Wang XY, Qin QY, Chen DF, Zhong QH, Wang L, et al. The prognostic role of BRAF mutation in metastatic colorectal cancer receiving anti-EGFR monoclonal antibodies: a meta-analysis. PLoS One. 2013;8(6):e65995. doi: 10.1371/journal.pone.0065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pai RK, Jayachandran P, Koong AC, Chang DT, Kwok S, Ma L, et al. BRAF-mutated, microsatellite-stable adenocarcinoma of the proximal colon: an aggressive adenocarcinoma with poor survival, mucinous differentiation, and adverse morphologic features. Am J Surg Pathol. 2012;36(5):744–52. doi: 10.1097/PAS.0b013e31824430d7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kakar S, Deng G, Sahai V, Matsuzaki K, Tanaka H, Miura S, et al. Clinicopathologic characteristics, CpG island methylator phenotype, and BRAF mutations in microsatellite-stable colorectal cancers without chromosomal instability. Arch Pathol Lab Med. 2008;132(6):958–64. doi: 10.5858/2008-132-958-CCCIMP. [DOI] [PubMed] [Google Scholar]
- 36.Ogino S, Nosho K, Kirkner GJ, Kawasaki T, Meyerhardt JA, Loda M, et al. CpG island methylator phenotype, microsatellite instability, BRAF mutation and clinical outcome in colon cancer. Gut. 2009;58(1):90–6. doi: 10.1136/gut.2008.155473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Samowitz WS, Sweeney C, Herrick J, Albertsen H, Levin TR, Murtaugh MA, et al. Poor survival associated with the BRAF V600E mutation in microsatellite-stable colon cancers. Cancer Res. 2005;65(14):6063–9. doi: 10.1158/0008-5472.CAN-05-0404. [DOI] [PubMed] [Google Scholar]
- 38.Goldstein J, Tran B, Ensor J, Gibbs P, Wong HL, Wong SF, et al. Multicenter retrospective analysis of metastatic colorectal cancer (CRC) with high-level microsatellite instability (MSI-H) Ann Oncol. 2014;25(5):1032–8. doi: 10.1093/annonc/mdu100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mao C, Zhou J, Yang Z, Huang Y, Wu X, Shen H, et al. KRAS, BRAF and PIK3CA mutations and the loss of PTEN expression in Chinese patients with colorectal cancer. PLoS One. 2012;7(5) doi: 10.1371/journal.pone.0036653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li HT, Lu YY, An YX, Wang X, Zhao QC. KRAS, BRAF and PIK3CA mutations in human colorectal cancer: relationship with metastatic colorectal cancer. Oncol Rep. 2011;25(6):1691–7. doi: 10.3892/or.2011.1217. [DOI] [PubMed] [Google Scholar]
- 41.Shen H, Yuan Y, Hu HG, Zhong X, Ye XX, Li MD, et al. Clinical significance of K-ras and BRAF mutations in Chinese colorectal cancer patients. World J Gastroenterol. 2011;17(6):809–16. doi: 10.3748/wjg.v17.i6.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Samowitz WS, Curtin K, Schaffer D, Robertson M, Leppert M, Slattery ML. Relationship of Ki-ras mutations in colon cancers to tumor location, stage, and survival: a population-based study. Cancer Epidemiol Biomarkers Prev. 2000;9(11):1193–7. [PubMed] [Google Scholar]
- 43.Andreyev HJ, Norman AR, Cunningham D, Oates J, Dix BR, Iacopetta BJ, et al. Kirsten ras mutations in patients with colorectal cancer: the ‘RASCAL II’ study. Br J Cancer. 2001;85(5):692–6. doi: 10.1054/bjoc.2001.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreyev HJ, Norman AR, Cunningham D, Oates JR, Clarke PA. Kirsten ras mutations in patients with colorectal cancer: the multicenter “RASCAL” study. J Natl Cancer Inst. 1998;90(9):675–84. doi: 10.1093/jnci/90.9.675. [DOI] [PubMed] [Google Scholar]
- 45.Consorti F, Lorenzotti A, Midiri G, Di Paola M. Prognostic significance of mucinous carcinoma of colon and rectum: a prospective case–control study. J Surg Oncol. 2000;73(2):70–4. doi: 10.1002/(SICI)1096-9098(200002)73:2<70::AID-JSO3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 46.Secco GB, Fardelli R, Campora E, Lapertosa G, Gentile R, Zoli S, et al. Primary mucinous adenocarcinomas and signet-ring cell carcinomas of colon and rectum. Oncology. 1994;51(1):30–4. doi: 10.1159/000227306. [DOI] [PubMed] [Google Scholar]
- 47.Blons H, Emile JF, Le Malicot K, Julie C, Zaanan A, Tabernero J, et al. Prognostic value of KRAS mutations in stage III colon cancer: post-hoc analysis of the PETACC8 phase III trial dataset. Ann Oncol. 2014;25(12):2378–85. doi: 10.1093/annonc/mdu464. [DOI] [PubMed] [Google Scholar]
- 48.Guerrero S, Casanova I, Farre L, Mazo A, Capella G, Mangues R. K-ras codon 12 mutation induces higher level of resistance to apoptosis and predisposition to anchorage-independent growth than codon 13 mutation or proto-oncogene overexpression. Cancer Res. 2000;60(23):6750–6. [PubMed] [Google Scholar]
- 49.Cespedes MV, Sancho FJ, Guerrero S, Parreno M, Casanova I, Pavon MA, et al. K-ras Asp12 mutant neither interacts with Raf, nor signals through Erk and is less tumorigenic than K-ras Val12. Carcinogenesis. 2006;27(11):2190–200. doi: 10.1093/carcin/bgl063. [DOI] [PubMed] [Google Scholar]
- 50.Malumbres M, Barbacid M. RAS oncogenes: the first 30 years. Nat Rev Cancer. 2003;3(6):459–65. doi: 10.1038/nrc1097. [DOI] [PubMed] [Google Scholar]
- 51.Ward RL, Todd AV, Santiago F, O’Connor T, Hawkins NJ. Activation of the K-ras oncogene in colorectal neoplasms is associated with decreased apoptosis. Cancer. 1997;79(6):1106–13. doi: 10.1002/(SICI)1097-0142(19970315)79:6<1106::AID-CNCR8>3.0.CO;2-D. [DOI] [PubMed] [Google Scholar]
- 52.Kumar SS, Price TJ, Mohyieldin O, Borg M, Townsend A, Hardingham JE. KRAS G13D mutation and sensitivity to cetuximab or panitumumab in a colorectal cancer cell line model. Gastrointest Cancer Res. 2014;7(1):23–6. [PMC free article] [PubMed] [Google Scholar]
- 53.Chen CC, Er TK, Liu YY, Hwang JK, Barrio MJ, Rodrigo M, et al. Computational analysis of KRAS mutations: implications for different effects on the KRAS p.G12D and p.G13D mutations. PLoS One. 2013;8(2):e55793. doi: 10.1371/journal.pone.0055793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douillard JY, Oliner KS, Siena S, Tabernero J, Burkes R, Barugel M, et al. Panitumumab-FOLFOX4 treatment and RAS mutations in colorectal cancer. N Engl J Med. 2013;369(11):1023–34. doi: 10.1056/NEJMoa1305275. [DOI] [PubMed] [Google Scholar]


