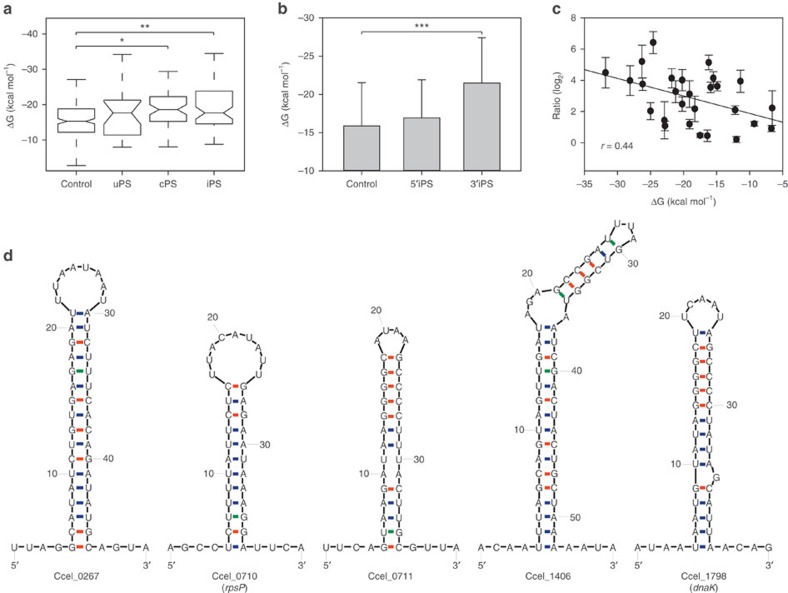
Figure 4. Mechanism underlying the distinct ratio in transcript abundance among the genes in ‘Complex' type operons.
(a) The RNA folding free energy (ΔG) of sequences flanking uPSs (N=24), cPSs (N=59) and iPSs (N=72). Sequences from non-PS intergenic regions within operons were used as the control (N=195). *P value=0.0005; **P value=0.0001 (Student's t-test). (b) Comparison of the average ΔG between the 5′iPS-proximal sequences (N=41) and the 3′iPS-proximal sequences (N=31). Sequences from non-PS intergenic region within operons were as the control. Error bars indicate s.d. of mean of all sequences in each type (***P value<0.0001, Student's t-test). (c) Correlation of the degree of transcript protection with folding free energy (ΔG) of the 3′iPS associated stem-loops. The degree of protection was quantified by the ratio in transcript abundance between the upstream and downstream genes that flank the iPSs. (d) The 3′iPS associated stem-loop structures that are, respectively, located at the 3′ end of Ccel_0267, rpsP, Ccel_0711, Ccel_1406 and dnaK. These structures are found in the operons shown in Fig. 3.