Figure 6.
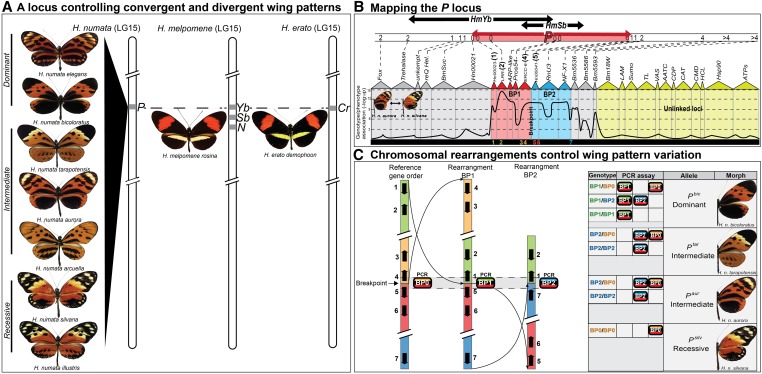
Characterizing the H. numata mimicry supergene. (A) A single Mendelian locus with multiple alleles (the P locus) controls wing-pattern diversity in H. numata. The P locus is positionally homologous to the Yb-Sb-N loci of H. melpomene and the Cr locus of H. erato (Joron et al. 2006b). (B) Fine-mapping and SNP associations narrow the P locus to a 400-kb interval spanning 31 genes and provide evidence of highly reduced recombination (red and blue areas) (Joron et al. 2011). Relative position of the genes (1–7) across the interval that were used to characterize genomic rearrangements is also shown. (C) Allelic variation at the P locus ultimately traces back to an inversion polymorphism with different wing-pattern morphs determined by distinct, nonrecombining haplotypes (Joron et al. 2011). PCR assay of the alternative breakpoints BP0, BP1, and BP2 (left) are perfectly associated with mimicry variation across four distinct morphs in eastern Peru (right).