Figure 4.
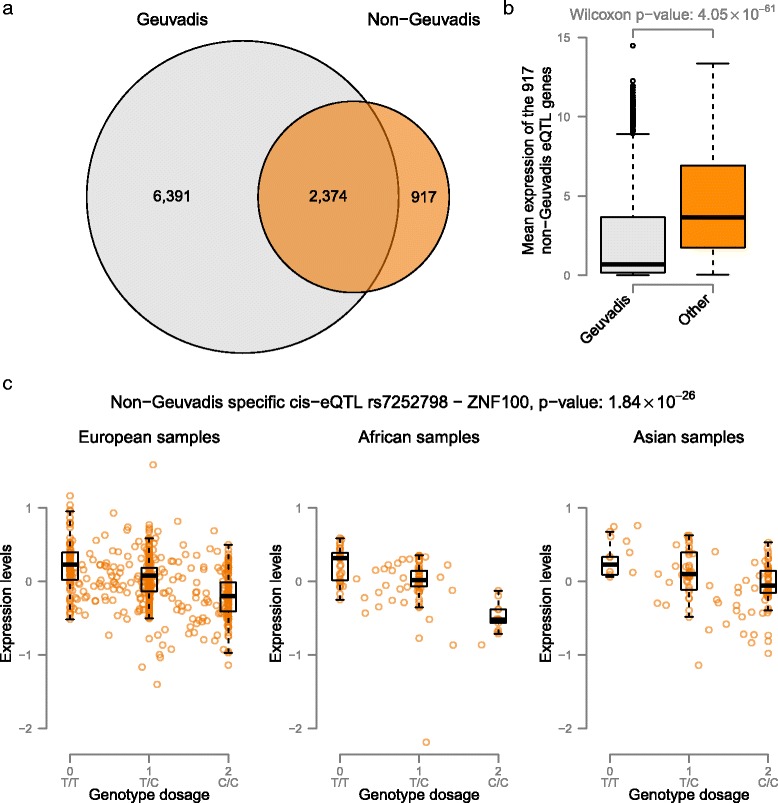
Geuvadis eQTLs versus non-Geuvadis eQTLs. (a) When performing eQTL analysis on the non-Geuvadis samples we identify 3,559 significant eQTL genes (FDR <0.05). Of these genes, 903 are not detected when using the Geuvadis samples. (b) Comparing the expression levels of the genes not identified in the Geuvadis samples we observe that, in general, these genes are much more abundantly expressed in the non-Geuvadis samples. (c) Example eQTL effect of rs7252798 affecting expression levels of ZNF100 that is only identified in the non-Geuvadis samples.
