Abstract
The past decade has witnessed steady and rapid progress in HCV research, which has led to the recent breakthrough in therapies against this significant human pathogen. Yet a deeper understanding of the life cycle of the virus is required to develop more affordable treatments and to advance vaccine design. HCV entry presents both a challenge for scientific research and an opportunity for alternative intervention approaches, owning to its highly complex nature and the myriad of players involved. More than half a dozen cellular proteins are implicated in HCV entry; and a more definitive picture regarding the structures of the glycoproteins is emerging. A role of apolipoproteins in HCV entry has also been established. Still, major questions remain, and the answers to these, which we summarize in this review, will hopefully close the gaps in our understanding and complete the puzzle that is HCV entry.
Keywords: apolipoproteins, cell polarity, entry receptors, glycoproteins, HCV, lipoviralparticles, membrane fusion, tight junction, very low-density lipoprotein particles, viral entry
HCV has been a major health burden in the past decades, infecting 130–170 million people worldwide. The virus produces chronic infection in the majority of patients and often remains undetected until late-stage symptoms emerge upwards of 20 years after infection. Without proper treatment, resulting conditions such as cirrhosis and eventual hepatocellular carcinoma can require extensive surgical remedy. Until recently, HCV had been difficult to treat, with highly varied treatment outcomes depending on many parameters, including the genotype of the virus (i.e., genotype 1 is less responsive to interferon/ribavirin therapy [1]) and the genetic background of the patients [2,3]. Newly approved therapies have come a long way in improving patient prognosis. The direct acting antiviral (DAA) combinations have brought about significant improvement in treatment outcome for genotype 1, with cure rates reaching greater than 90% without the need for interferon or ribavirin (reviewed in [4]). However, the substantial cost of these DAAs prevents their broad distribution; and an even bigger concern is the small percentage of infected individuals receiving treatment. Most patients do not receive diagnosis until many years after infection, and treatment alone may not be sufficient to systematically reach all patients, or be adequate to protect against new infections. As such, a deeper understanding of HCV biology is needed in order to guide development of more affordable treatment options. In addition, a prophylactic vaccine is likely still ultimately required to eradicate HCV infections globally, and comprehensive knowledge of HCV entry molecules and pathways will greatly facilitate the design of vaccine candidates.
The virus family Flaviviridae consists of the genera Flavivirus, Pegivirus, Pestivirus and the HCV-containing Hepacivirus. The virions of this virus family all contain a RNA genome that is encased in a viral protein structure called the capsid. This structure is in turn enveloped in a host-derived membrane. Additionally, all members of the family share a similar genome organization and mechanism of replication.
HCV has a positive-sense RNA genome of 9.6 kb in length. Upon entering human hepatocytes, the genome is used to as a template to produce a single polypeptide which is further cleaved by host and viral proteases into 10 proteins. Six of these proteins are designated as nonstructural proteins: NS2, NS3, NS4A, NS4B, NS5A and NS5B and function inside the host cell to replicate the viral genome. Although none of these proteins, or the ion channel protein P7, is incorporated into virions, many of them nevertheless act to facilitate the assembly of new viral particles for subsequent rounds of infection. There has been great success in development of antivirals targeting the nonstructural proteins; the newest DAA therapies consist of inhibitors targeting the RNA-dependent RNA polymerase (NS5B), the multifunctional phosphoprotein (NS5A; reviewed in [5]) and the serine protease (NS3/4A; reviewed in [6]).
The three structural proteins (Core, E1 and E2) are the physical components of the HCV virion. The core protein forms the capsid, encasing the RNA genome. The capsid is then surrounded by a host-derived membrane studded with the viral glycoproteins, E1 and E2. In conjunction with host proteins, the glycoproteins facilitate the attachment and eventual internalization of the viral particle. The arrangement of E1 and E2 of the surface of the virion were initially predicted to be similar to that of flaviviruses [7–10], however more recent ultrastructural imaging has raised question on this idea [11]. E1 and E2 form a stable heterodimer via interactions at their transmembrane domains, and it is currently unclear which of the glycoproteins acts as the fusion protein [12–18].
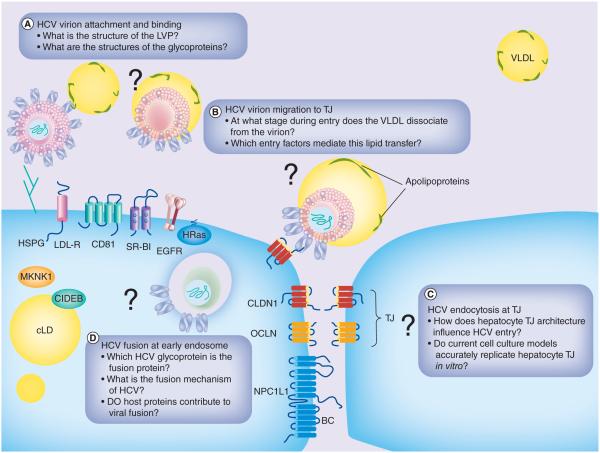
The entry process for HCV consists of several sequential steps. The virus must first attach to cells, via interactions with cell surface host proteins, and eventually binding to surface receptors that guide the virion to the tight junction (TJ). From there, the virion is internalized via clathrin-mediated endocytosis and the virion is maintained in the early endosome until fusion. The HCV research community has made great strides in generating a significant amount of knowledge regarding entry, and several outstanding reviews can be found in the literature (see [19–22]). In this review, we will aim to discuss several active areas of research in HCV entry and to pose questions that address the remaining missing pieces of the HCV-entry puzzle (Figure 1).
Figure 1. Unresolved questions at various stages of HCV entry.
BC: Bile canaliculus; cLD: Cytoplasmic lipid droplet; EGFR: EGF receptor; HSPG: Heparan sulfate proteoglycans; LDL-R: Low-density lipoprotein receptor; LVP: Lipoviralparticle; TJ: Tight junction; VLDL: Very low-density lipoprotein.
Pieces of all shapes & sizes: lipoviralparticle and HCV entry
Very low-density lipoproteins
Understanding the physical attributes of the infectious particles is important for HCV entry research. A major feature of HCV virions is their association with lipoproteins [19,23], which are normally involved in the release and uptake of lipid species in the liver [24]. Indeed, this unusual aspect of HCV virion structure is an important part of the close relationship between HCV infection and liver lipid metabolism [25].
Hepatocytes regulate the intake and efflux of lipids from the cell via lipoprotein particles. These apolipoprotein-associated particles are termed high density lipoprotein (HDL), low-density lipoprotein (LDL), very low-density lipoprotein (VLDL) and chylomicron particles based on their various buoyant densities [19,23,24]. Apolipoproteins, such as apoB-100 and its truncated form apoB-48, are associated with the lipid monolayer surrounding the TAG core of VLDLs [26,27].
Secreted VLDLs range in size from 25 to 80 nm in diameter [28]. Formation of the immature VLDLs starts in the lumen of the ER where initial lipidation is facilitated by microsomal triglyceride transfer protein (MTP) and the presence of apoB. Each VLDL particle contains a single copy of apoB-100 and exchangeable amounts of apoC and apoE [29–31]. In addition to the VLDL produced from liver cells, intestinal cells also secrete similar particles, known as chylomicrons, decorated with a single copy of apoB-48 [32–34].
Close association of lipoproteins with HCV makes for the lipoviralparticle
HCV virions are believed to associate with VLDLs [11,35–38] and as a result display a lower density compared with related enveloped RNA viruses (for comparison, see [19]). The buoyant density of HCV particles also varies significantly and is related to specific infectivity of the particles [39–42]. HCV virions, either derived from patient sera or produced in cell culture (HCVcc), are not uniformly infectious; in fact, ratios varying from 1:100 to 1:1000 of infectious to subviral particles have been observed [43–46]. The particles with the buoyant densities between 1.09–1.10 g/ml are the most infectious [44,47], while the other particles either demonstrate lower infectivity or contain no viral RNA despite expression of HCV glycoproteins on the viral envelope surface [48,49].
The association of HCV particles and VLDLs leads to the formation of the lipoviralparticles (LVP) which are believed to be the functional units of HCV infection [23,50]. In addition to the relatively low buoyant density, evidence for LVPs comes from the detection of many lipoproteins, such as apoA-1, apoB-100 or apoB-48, apoC-1 and apoE, in purified virion preparations [26,27,51,52]. While all these apolipoproteins have been found to associate with serum-derived particles, the extent of association is more limited in HCVcc. For example, apoB-100 is readily found on serum-derived LVPs, the presence of apoB-100 on HCVcc virions is less clear [26,37,49,50,52,53]; and apoE and apoC-I appear to be the major observable apolipoprotein species [36,37,54] on the HCVcc particles. In addition to the difference between HCV serum and HCVcc, the range of LVP buoyant densities also varies depending upon the host or cell type used for the production of particles [48,49,53,55]. This brings into question if the current cell-culture systems, such as Huh7.5, are ideal models for analyzing the importance of LVP.
LVP structure has not been determined, however, ultrastructure images of purified immunolabeled virions have revealed the diversity of LVP shapes and sizes [11,35,56]. One model of LVP structure proposes that the VLDL TAG contents are incorporated in-between the leaflets of the viral envelope [23,50]. This hypothesis is supported by the observation that virion assembly depends on VLDL maturation, suggesting LVP formation may utilize the VLDL maturation pathway [57,58]. A second model suggests LVPs may consist of two connected particles (viral and VLDL), bridged by apolipoproteins [59]. This idea is supported by the purification of HCV virions associated with apoB-48 containing chylomicrons [52]. However, at what stage during virion production the association with VLDL occurs is unclear. While the specifics of LVP structure remain unknown, the LVP components likely influence the uptake of virions in new rounds of infection.
Impact of VLDL association on HCV entry
It has been estimated that there are approximately 300 copies of apoE per infectious HCV virion [35,60] and apoE likely plays a dual role in the assembly of infectious particles as well as in modulating HCV entry. Heparan sulfate proteoglycans (HSPGs) have been shown to facilitate HCV attachment through interaction with apoE and this binding has been demonstrated for both genotypes 2A and 1B virions. Addition of HSPG-binding peptides derived from apoE sequence prevented virions from binding to heparan in a dose-dependent manner, and deletion of HSPG from the surface of liver cells suppressed virion attachment [61–64].
SR-BI interaction with both apoB and apoE has been proposed to facilitate its function in HCV entry. Chinese hamster ovary (CHO) cells expressing surface SR-BI were capable of mediating binding of patient serum-derived HCV particles and this interaction was insensitive to anti-E2 antibodies. The interaction was however outcompeted by apoB-containing particles, predominantly VLDL, implicating apoB in the binding of the HCV particles [65]. Deletion of HCV E2 HVR1 reduced virion dependence on apoE for attachment [66], suggesting this attachment step may involve the interactions of both apoE and E2 with SR-BI. In vivo experiments have also demonstrated a role for SR-BI in VLDL uptake, as SR-BI−/− mice exhibited elevated serum VLDL and a reduced association of VLDL with hepatocytes [67], again arguing for a general role of SR-BI in the attachment of lipoproteins such as those associated with HCV LVPs.
Hishiki et al. examined the role of each of three apoE isoforms in modulating HCV virion dependence on low-density lipoprotein receptor (LDL-R) or SR-BI for entry. SiRNA-mediated knockdown of LDL-R or SR-BI reduced infection of HCV virions expressing apoE3. However, this effect was not observed when using HCV virions expressing apoE2, a smaller isoform of apoE that exhibits lower affinity for LDL-R and SR-BI [68], suggesting the observed reduction in infection for HCV/apoE3 was dependent on interactions with LDL-R or SR-BI. Despite this and another report of correlation between the dependence on LDL-R for entry and apoE-containing particles [69], the role of LDL-R in apoE-mediated HCV entry is controversial [70].
Sorting out the pieces: HCV entry factors
All viruses depend on host cell receptors for entry to target cells. A large number of cellular factors have been identified to be involved in HCV entry. An general outline of the discrete steps of the entry process and the protein involved in these step have been illustrated (Figure 1). In this section, we will discuss how each of these proteins was identified and what roles that they play in the entry process.
Entry factors identified via binding to HCV glycoproteins
A common approach to discover viral entry receptors is to find host cell-surface proteins that directly interact with the viral glycoproteins. For HCV, several entry factors were identified using this approach. CD81, a critical HCV receptor, was identified as a cell surface protein that binds to a soluble peptide of HCV E2 in a cell-based screen [71]. The interaction was later confirmed using HIV/HCV pseudoparticles (HCVpp) expressing the HCV glycoproteins [72–74] . Blocking CD81 function in vitro with CD81 antibodies, soluble CD81, or siRNA all inhibited HCV entry [44,73,75–80]. The importance of CD81 function in HCV life cycle was further highlighted by the ability of an CD81 monoclonal antibody to suppress HCV infection in vivo [81,82]. CD81 contains two extracellular loops termed the small extracellular loop (SEL) and the large extracellular loop (LEL) [83]. The LEL is noted for containing a CCG motif which allows the formation of disulfide bridges found to be critical for interaction with E2 [71,74,84–86]. The interaction of CD81 and HCV E2 is an essential early step during HCV entry and likely required for the translocation of the virion/receptor complex to the TJs.
Another indispensable HCV receptor is SR-BI. It was also identified based on its interaction with E2. HepG2 cells were incubated with a tagged soluble E2 and the interacting receptors were purified by co-immunoprecipitation after crosslinking. The major E2-binding protein was identified to be SR-BI. The interaction site was further mapped to the hyper variable region 1 (HRV1) of HCV E2 [87]. The importance of SR-BI in HCV entry has been confirmed by numerous in vitro and in vivo studies [80,87–93]. SR-BI may also facilitate additional aspects of HCV attachment independent of glycoprotein binding [93].
Additional E2-binding proteins have been implicated in HCV infection. These include DC-SIGN, DC-SIGNR [94] , which may deliver HCV particles in the bloodstream from the sinusoids to hepatocytes [95]. And the HSPGs have been suggested to directly interact with HCV E2 [96,97], in addition to the well-established HSPG-apolipoprotein interactions [36,61,62,98]. Several E1-binding proteins have also been reported [99–101], but the role of E1 in receptor-binding is not understood.
Entry factors identified through cDNA functional complementation
Another important approach for receptor discovery is cDNA screening for genes that can confer permissiveness for viral entry onto nonpermissive cells. The identification of a TJ protein, CLDN1, as a HCV entry cofactor, was accomplished with such a cDNA complementation assay, using nonpermissive cells expressing CD81 and SR-BI and cDNA library constructed from permissive cells [102]. CLDN1 interacts with CD81 and acts during a postbinding step, likely at the hepatocyte TJ, where the virions migrate to before internalization [103–105]. PKA signaling is important for this interaction, as inhibition of PKA signaling results in disruption of CD81-CLDN1 interactions and altered localization of CLDN1 [106]. A recent study suggest that E1/E2 dimer may also interact with CLDN1 [107]. Related molecules CLDN6 and CLDN9 have also been suggested to act as HCV entry receptors [108–110], but while CLDN1 supports infection of a broad range of HCV isolates in Huh6 cells, CLDN6 only supports infection by a limited number of isolates [110]. Additionally, work by Fofana et al. demonstrated that the expression of CLDN6 and CLDN9 were very low in primary human hepatocytes (PHHs) and that CLDN6- and CLDN9-specific antibodies fail to restrict HCVpp entry into these primary cells [111].
Another TJ protein, OCLN, was identified as an HCV entry cofactor by two independent approaches, siRNA knockdown testing [112] and cDNA complementation [80]. The mouse NIH3T3 cells engineered to express the other three human receptors, CD81, SR-BI and CLDN1 were used as nonpermissive cells in the expression cloning this time [80]. These four molecules together constitute the full complement of human proteins that can enable HCV entry into mouse cells. As a result, their discovery enabled the establishment of immunocompetent mouse models of HCV entry and infection [82,113].
Entry factors identified through siRNA library screens
Screening of siRNA libraries, either genomewide [114] or targeted at a specific subset of genes [115,116], has been successfully applied toward the discovery of cellular cofactors of HCV entry. EGFR and EphA2 were both found to be important for HCV entry through a human kinome siRNA screen [115]. EGFR was shown to promote the CD81-CLDN1 interaction and EGFR was internalized in an CD81-dependent manner upon infection [117]. This effect is likely mediated by a membrane-bound GTPase, HRas, which is involved in the EGFR signaling pathway. HRas signaling was required for CD81 lateral membrane diffusion, which is important for CD81-CLDN1 interaction and migration of the virion to the TJs [118]. Furthermore, a genome-wide siRNA library screen led to the identification of several additional host proteins that play a role in HCV entry. These include E-cadherin (CDH1), the Rho GTPase RAC1, a choline kinase (CHKA), the SMAD family member SMAD6 and a subunit of NADPH oxidase (CYBA) [114].
Other approaches
Based on the observation that cholesterol plays a critical role in HCV entry [91,119], Sainz et al. scrutinized the role of the cholesterol receptor NPC1L1 as a potential HCV entry factor [120]. NPC1L1 is an integral membrane molecule found on the apical membrane of hepatocytes. Its localization in the bile canaliculus facilitates the uptake of cholesterol into the surrounding hepatocytes via clathrin-mediated endocytosis [121]. Knockdown of NPC1L1 inhibited HCV infection, and blocking NPC1L1 with the inhibitor ezetimibe suppressed pangenotypic HCV infections in vitro [120] . The mechanism by which NPC1L1 mediates entry is still unknown, but as HCV particles enter predominantly through the basolateral membrane, it is possible that NPC1L1 is acting at a step postinternalization, such as the removal of cholesterol from the incoming virions as HCV virions are enriched in cholesterol content [119,122]. The infectivity of HCV particles appears to be highly sensitive to modulations of virion-associated cholesterol levels. On one hand, depletion of cholesterol from HCVcc virions with methyl-β-cyclodextrin inhibited virion internalization, a defect that could be rescued by supplying exogenous cholesterol [119]. On the other hand, when HCVpp containing different levels of cholesterol were tested for sensitivity to a NPC1L1 inhibitor, only the HCVpp particles with higher cholesterol content were shown to be dependent on NPC1L1 [120], suggesting a role of NPC1L1 in modulating cholesterol level during entry. Interestingly, the related protein NPC1 has previously been identified as an entry factor for ebola virus [123,124].
Analysis of differential gene expression between permissive and nonpermissive cells can provide insight into host determinants of infection. Utilizing an embryonic stem cell differentiation protocol to generate differentiated human hepatocyte-like cells, Wu et al. identified a discrete transition to HCV permissiveness during hepatic differentiation [125], which enabled gene profiling that revealed a liver-enriched protein important for HCV entry. Both knockdown with siRNA and knockout with TAL effector nuclease (TALEN) of this protein, CIDEB, inhibited entry of HCV, but not that of West Nile virus or vesicular stomatitis virus [126].
Screening of kinase inhibitors led to the observation that the MKNK1 plays a role in HCV infection. MKNK1 siRNA reduced HCV entry without affecting viral expression and RNA replication [127]. Other molecules that play a putative role in HCV attachment and entry include glycoaminoglycans (GAGs) [97], LDL-R [128] and the iron receptor TfR1 [129].
Keeping the pieces in place: role of the tight junction in entry
Following the CD81-E2 engagement, the HCV virion laterally diffuses to the TJ. As previously discussed, CLDN1 and OCLN, two integral TJ proteins, are essential for HCV entry; however, how the TJ architecture itself impacts virion internalization remains an area with many open questions.
Tight junctions define hepatocyte architecture
Many types of epithelial cells harbor various junctions which regulate tissue architecture and prevent the lateral diffusion of surface proteins between the basal and apical membranes, facilitating compartmentalization of the extracellular environment. Hepatocyte TJs separate blood and bile interaction as adjacent apical domains encase the bile canaliculi while the basolateral domains interact with the blood-containing sinusoids [130].
The claudin family of integral-membrane proteins maintains much of the integrity of the hepatocyte TJ. The claudin proteins on adjacent cells interact with each other via their extracellular loops to form the barrier [131]. Additionally, occludin, tricellulin and members of the immunoglobulin super family, such as JAM-A, JAM4, CAR and ESAM, make up the extracellular junction.
How important is tight junction architecture to HCV entry?
Although several TJ proteins have been identified as HCV entry receptors and HCV infection is capable of disrupting TJ in polarized cells [132–134], the role of the TJ architecture itself in HCV infection is less clear. For example, CLDN1 can to a lesser extent localize to the basolateral membrane in polarized cells [131], thus in theory can mediate infection independent of its role in TJ maintenance. The commonly-used Huh7.5 cells are not polarized [135] and it has been suggested that polarization may affect lipoprotein profiles. For example, a correlations between lipoprotein production and cell polarity in Caco-2 cells have been reported [136,137]. Consequently Huh7.5 cells may not recapitulate the TJ architecture of hepatocytes in vivo and may additionally produce altered lipid profiles. Indeed, LVP produced in nonpolarized cells have been shown to lack many of the associated lipoproteins seen in patient-derived virions [11,26,35–38,50,52]. All these could have contributed to the conflicting data surrounding the role of LDL-R in HCV uptake and the differences observed in entry efficiency between HCVcc and HCVpp [138–141].
The HepG2 cell line exhibits polarization [142] similar to cells surrounding bile canaliculi and have been modified with exogenous expression of miR-122 and CD81 to increase permissiveness to HCV infection [143]. Other laboratories have attempted to culture Huh7-based cells in alternative systems in order to simulate 3D cell contacts [144,145].
PHHs isolated from patient biopsies have been used to study HCV infection in vitro [53,146–152]. However, these cells suffer from limited accessibility, variability between batches, a finite culture time and low levels of infection with the standard HCVcc strains. Recently, several groups have reported differentiated human hepatocyte-like cells in vitro that are permissive for HCVcc infection [125,153,154]. These stem cell-derived cells are a renewable alternative to PHHs, however, the differentiation protocols are complex and the hepatocyte-like cells may not truly represent mature hepatocytes. As both cellular polarization and lipidation of virions likely contribute to the entry mechanism, additional in vitro models that combine sufficient hepatic features and three-dimensional engineering may be needed to fully understand the role of TJs in HCV entry.
Fitting the pieces together: the mechanism of HCV fusion
HCV entry culminates with the fusion of viral and host endosomal membranes in order to release the virion into the cytoplasm of the target cell. In contrast to many other positive strand RNA viruses, HCV fusion and uncoating steps are much less understood. One of the biggest challenges is the identification of the fusion protein and elucidation of the membrane fusion process that releases HCV capsid from the endosomes.
Alpha- & flavivirus fusion
HCV and related viruses predominantly enter target cells through clathrin-dependent endocytosis [75,155–157]. Once inside the endosomes, fusion between the viral membrane and the endosomal membrane is required to release the RNA-containing capsid. The fusion process is facilitated by the integral glycoproteins embedded in the virion membrane. The fusion proteins can be divided into three main classes based on their structures and the fusion mechanisms [158]. The alpha- and flavivirus fusion proteins both fall under class II, and there has been speculation as to whether HCV glycoproteins also fall into this class [158–160].
Much of the alpha- and flavivirus fusion mechanisms have been elucidated. Several crystal structures exist for flavivirus fusion protein E [161–164], and for the E1 glycoprotein of alphaviruses, which is also the fusion protein [165–168]. While the sequences of the fusion proteins vary signif icantly between these viruses, the 3D structural features are very similar. In addition to the fusion protein E1, alphaviruses encode another glycoprotein E2 which acts as the companion molecule that shields the fusion peptide of E1 from exposure prior to low pH-activated fusion. Flaviviruses also encode two glycoproteins, the E protein and the prM/M protein (companion protein). The prM protein undergoes a cleavage event by the furin protease in the trans-Golgi network during virion maturation to produce infectious particles. The mature M protein then remains in association with the E protein until activation of fusion [158].
New insights from a pestivirus
Pestiviruses also enter cells via glycoprotein-mediated membrane fusion. The pestivirus membrane is decorated with its own E1 and E2 glycoproteins [157]. The crystal structure of BVDV-1 E2 [169,170] revealed that the protein did not contain a class II fusion fold as originally predicted by computational analysis of sequence similarities [159], and that at low pH the structure was disordered [169,170]. The overall fold observed from crystal structure of BVDV-1 E2 does not show the three domain structure typical of alphaor flavivirus class II fusion folds. Instead, BVDV-1 E2 adopts an extended conformation of four linearly organized domains and presumably represents a novel class of fusion proteins [169]. In addition, there is another glycoprotein loosely bound to the membrane of the BVDV-1 virion. This heavily glycosylated protein is tethered to the virion by an amphipathic helix and can also be found in a secreted form. It has been shown to possess RNase activity, which may function to suppress viral RNA-induced type I interferon synthesis [171–173].
The elusive HCV fusion protein
In contrast to the alpha- and flaviviruses, the identity of the HCV fusion protein is unknown. Both of the HCV glycoproteins have been proposed to harbor putative fusion peptides [159,169,174–177], and initial HCVpp studies suggested exposure of new domains of E2 at low pH, leading to the prediction of E2 as a class II fusion protein [178], although it was already noted that the organization of the HCV E1 and E2 heterodimer differs significantly from those of the dimer and homotrimer formation of the dengue virus E protein. Because pestivirus fusion protein also acts differently from class II fusion proteins, an analogy was also drawn between bovine viral diarrhea virus 1 (BVDV-1) E2 and HCV E2 [157,159,169]. However, the recently solved crystal structure of the HCV E2 core ectodomain [16,17] suggests that these early predictions are not correct. Unlike the extended conformations of either flavivirus E protein or the BVDV-1 E2 protein, the HCV E2 ectodomain is compact and globular, with no significant change induced by lower pH [16,17]. These results indicate that either HCV E2 acts through a novel mechanism, or HCV E1 plays a greater role in inducing membrane fusion than previously thought. Both E1 and E2 are much smaller in comparison to known class II fusion proteins and the compact nature of E2 structure brings into question how the protein would be capable of spanning the full membrane. A partial crystal structure of HCV E1 amino acids 1-79 is now available [18] and shows no evidence of a fusion fold, but that may be because the structure only covers part of the putative fusion peptide [18,169,174]. A complete crystal structure of HCV E1 will shed light on its role in the fusion mechanism, and ultimately a structure of the E1/E2 heterodimer may be necessary to fully understand the interplay of these two proteins in inducing membrane fusion.
Conclusion & future perspective
The HCV field has made great strides in understanding the receptors facilitating the initial attachment and entry. Nevertheless, much research is still needed before understanding of the entry mechanism can be used to help guide vaccine design. The most imperative is determining the full crystal structures of the E1 and E2 glycoproteins, both pre- and post-fusion. The latest breakthrough on the structure of E2 ecto-domain has brought up as many questions as it answers by suggesting that HCV fusion occurs through a mechanism that is distinct from those of either flavivirus or pestivirus [16,17]. Progress in this regard likely requires overcoming the effect of heavy glycosylation of the E1 and E2 proteins on crystallization and modeling experiments. In addition, the structure of the LVP itself presents another challenge as the complex interactions of the lipoproteins have hindered efforts to determine glycoprotein structure displayed on virions using cryo-EM techniques [11,35,56].
A significant question about the nature of the LVP is the influence of lipid and lipoprotein association on the enhancement of HCV entry. Indeed, several HCV-binding and entry receptors, such as GAGs, SR-BI and possibly LDL-R, are normally involved in the uptake of lipoproteins. For example, these entry factors have been implicated in enhancing HCV infection following consumption of a fatty meal through rapid reuptake of newly produced LVP [26]. Additionally, the point in entry where the virion-associated lipids and lipoproteins are removed remains to be determined. These questions emphasize the importance of using viral particles produced in physiologically relevant systems for studying HCV entry. Such a system will need to simultaneously maintain cell polarity and produce virions more closely mimicking the lipoprotein content of those found in vivo. In conclusion, while new pieces of the HCV-entry puzzle are beginning to fall into place, there are still many questions remaining about the nature of HCV particles and how the combinations of viral and host proteins at the cell surface contribute to viral entry. With a clearer picture in mind, further work can focus on understanding the fusion and uncoating mechanism to finally piece together the HCV-entry puzzle.
EXECUTIVE SUMMARY.
HCV entry is mediated by many host factors
- These host factors and key receptors have been identified through multiple approaches:
- - Use of HCV glycoprotein binding assays, cDNA complementation assays, siRNA library screening and differential gene profiling have all proven successful in identifying HCV entry-related host factors;
- - CD81, SR-BI, CLDN and OCLN constitute the full complement of human proteins that can enable HCV entry into mouse cells.
The HCV fusion mechanism is unknown
The identity of the HCV fusion protein has not been determined, and the crystal structure of E2 ectodomain does not support predictions for a class II fusion peptide, such as those observed in alpha- and flavivirus fusion proteins.
The nature of the lipoviralparticle influences HCV entry
- HCV particles associate with VLDL and various apolipoproteins in vivo:
- - The association of HCV particles and VLDLs leads to the formation of the lipoviralparticles (LVP) which are believed to be the functional units of HCV infection;
- - Isolated LVP has been found in association with various apolipoproteins, such as apoE, apoB-100 or apoB-48, apoA-1 and apoC-1.
These associations contribute to HCV’s low buoyant density relative to other RNA viruses. HCV particles are diverse in shape and size, with lower buoyant density corresponding to higher specific infectivity.
Tight junction architecture needs to be considered when studying HCV entry
- The critical HCV entry receptors, CLDN1 and OCLN, are both tight junction proteins:
- - The incoming HCV virion migrates to the TJ after initial attachment and binding to the receptor CD81 on the basolateral membrane.
The most common HCV cell culture systems lack typical hepatocyte polarization, and future studies need to consider additional models for understanding the role of liver architecture in HCV entry.
Acknowledgements
The authors apologize to all their colleagues whose important work could not be directly cited.
Related research in the authors’ lab is supported by NIH grants R21AI111250 and R56AI107763 to H Tang.
Footnotes
Financial & competing interests disclosure
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as:
• of interest; •• of considerable interest
- 1.Martinot-Peignoux M, Marcellin P, Pouteau M, et al. Pretreatment serum hepatitis C virus RNA levels and hepatitis C virus genotype are the main and independent prognostic factors of sustained response to interferon alfa therapy in chronic hepatitis C. Hepatology. 1995;22(4):1050–6. Pt 1. [PubMed] [Google Scholar]
- 2.Ge D, Fellay J, Thompson AJ, et al. Genetic variation in IL28B predicts hepatitis C treatment-induced viral clearance. Nature. 2009;461:399–401. doi: 10.1038/nature08309. [DOI] [PubMed] [Google Scholar]
- 3.Thomas DL, Thio CL, Martin MP, et al. Genetic variation in IL28B and spontaneous clearance of hepatitis C virus. Nature. 2009;461:798–801. doi: 10.1038/nature08463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kohli A, Shaffer A, Sherman A, Kottilil S. Treatment of hepatitis C. JAMA. 2014;312(6):631. doi: 10.1001/jama.2014.7085. [DOI] [PubMed] [Google Scholar]
- 5.Paul D, Madan V, Bartenschlager R. Hepatitis C virus RNA replication and assembly: living on the fat of the land. Cell Host Microbe. 2014;16(5):569–579. doi: 10.1016/j.chom.2014.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hayes CNN, Chayama K. Emerging treatments for chronic hepatitis C. J. Formos. Med. Assoc. 2014 doi: 10.1016/j.jfma.2014.09.001. [DOI] [PubMed] [Google Scholar]
- 7.Kuhn RJ, Zhang W, Rossmann MG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mukhopadhyay S, Kim B-S, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302(5643):248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 9.Yu X, Qiao M, Atanasov I, et al. Cryo-electron microscopy and three-dimensional reconstructions of hepatitis C virus particles. Virology. 2007;367:126–134. doi: 10.1016/j.virol.2007.05.038. [DOI] [PubMed] [Google Scholar]
- 10.Bonnafous P, Perrault M, Le Bihan O, et al. Characterization of hepatitis C virus pseudoparticles by cryo-transmission electron microscopy using functionalized magnetic nanobeads. J. Gen. Virol. 2010;91:1919–1930. doi: 10.1099/vir.0.021071-0. [DOI] [PubMed] [Google Scholar]
- 11.Catanese MT, Uryu K, Kopp M, et al. Ultrastructural analysis of hepatitis C virus particles. Proc. Natl Acad. Sci. USA. 2013;110:9505–10. doi: 10.1073/pnas.1307527110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vieyres G, Thomas X, Descamps V, Duverlie G, Patel AH, Dubuisson J. Characterization of the envelope glycoproteins associated with infectious hepatitis C virus. J. Virol. 2010;84:10159–10168. doi: 10.1128/JVI.01180-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dubuisson J, Hsu HH, Cheung RC, Greenberg HB, Russell DG, Rice CM. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J. Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Duvet S, Cocquerel L, Pillez A, et al. Hepatitis C virus glycoprotein complex localization in the endoplasmic reticulum involves a determinant for retention and not retrieval. J. Biol. Chem. 1999;273:32088–32095. doi: 10.1074/jbc.273.48.32088. [DOI] [PubMed] [Google Scholar]
- 15.Rouillé Y, Helle F, Delgrange D, et al. Subcellular localization of hepatitis C virus structural proteins in a cell culture system that efficiently replicates the virus. J. Virol. 2006;80:2832–2841. doi: 10.1128/JVI.80.6.2832-2841.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kong L, Giang E, Nieusma T, et al. Hepatitis C virus E2 envelope glycoprotein core structure. Science. 2013;342:1090–4. doi: 10.1126/science.1243876. •• Presents the first crystal structure of the HCV E2 ectodomain, revealing novel aspects of the glycoprotein. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Khan AG, Whidby J, Miller MT, et al. Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2. Nature. 2014;509(7500):381–4. doi: 10.1038/nature13117. •• Confirms the globular nature of the HCV E2 crystal structure. Together with the reference above, these works reveal unexpected aspects of the HCV E2 glycoprotein structure which can be used to guide vaccine design. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.El Omari K, Iourin O, Kadlec J, et al. Unexpected structure for the N-terminal domain of hepatitis C virus envelope glycoprotein E1. Nat. Commun. 2014;5:4874. doi: 10.1038/ncomms5874. • Provides the first insights into the structure of HCV E1 through a successful crystalization of amino acids 1–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lindenbach BD, Rice CM. The ins and outs of hepatitis C virus entry and assembly. Nat. Rev. Microbiol. 2013;11(10):688–700. doi: 10.1038/nrmicro3098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Baumert TF, Meredith L, Ni Y, Felmlee DJ, McKeating JA, Urban S. Entry of hepatitis B and C viruses - recent progress and future impact. Curr. Opin. Virol. 2014;4:58–65. doi: 10.1016/j.coviro.2013.12.002. [DOI] [PubMed] [Google Scholar]
- 21.Zeisel MB, Fofana I, Fafi-Kremer S, Baumert TF. Hepatitis C virus entry into hepatocytes: molecular mechanisms and targets for antiviral therapies. J. Hepatol. 2011;54:566–576. doi: 10.1016/j.jhep.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 22.Ding Q, von Schaewen M, Ploss A. The impact of hepatitis C virus entry on viral tropism. Cell Host Microbe. 2014;16(5):562–568. doi: 10.1016/j.chom.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartenschlager R, Penin F, Lohmann V, André P. Assembly of infectious hepatitis C virus particles. Trends Microbiol. 2011;19:95–103. doi: 10.1016/j.tim.2010.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Bassendine MF, Sheridan DA, Felmlee DJ, Bridge SH, Toms GL, Neely RDG. HCV and the hepatic lipid pathway as a potential treatment target. J. Hepatol. 2011;55:1428–1440. doi: 10.1016/j.jhep.2011.06.004. [DOI] [PubMed] [Google Scholar]
- 25.Felmlee DJ, Hafirassou ML, Lefevre M, Baumert TF, Schuster C. Hepatitis C virus, cholesterol and lipoproteins--impact for the viral life cycle and pathogenesis of liver disease. Viruses. 2013;5:1292–324. doi: 10.3390/v5051292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Felmlee DJ, Sheridan DA, Bridge SH, et al. Intravascular transfer contributes to postprandial increase in numbers of very-low-density hepatitis C virus particles. Gastroenterology. 2010;139:1774–1783. doi: 10.1053/j.gastro.2010.07.047. [DOI] [PubMed] [Google Scholar]
- 27.Thomssen R, Bonk S, Propfe C, et al. Association of hepatitis C virus in human sera with beta-lipoprotein. Med. Microbiol. Immunol. 1992;181:293–300. doi: 10.1007/BF00198849. [DOI] [PubMed] [Google Scholar]
- 28.Alexander CA, Hamilton RL, Havel RJ. Subcellular localization of B apoprotein of plasma lipoproteins in rat liver. J. Cell Biol. 1976;69:241–263. doi: 10.1083/jcb.69.2.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elovson J, Chatterton JE, Bell GT, et al. Plasma very low density lipoproteins contain a single molecule of apolipoprotein B. J. Lipid Res. 1988;29:1461–1473. [PubMed] [Google Scholar]
- 30.Cohn JS, Tremblay M, Batal R, et al. Plasma kinetics of VLDL and HDL apoC-I in normolipidemic and hypertriglyceridemic subjects. J. Lipid Res. 2002;43:1680–1687. doi: 10.1194/jlr.m200055-jlr200. [DOI] [PubMed] [Google Scholar]
- 31.Mahley RW. Apolipoprotein E: cholesterol transport protein with expanding role in cell biology. Science. 1988;240:622–630. doi: 10.1126/science.3283935. [DOI] [PubMed] [Google Scholar]
- 32.Hussain MM, Rava P, Pan X, et al. Microsomal triglyceride transfer protein in plasma and cellular lipid metabolism. Curr. Opin. Lipidol. 2008;19:277–284. doi: 10.1097/MOL.0b013e3282feea85. [DOI] [PubMed] [Google Scholar]
- 33.Rutledge AC, Su Q, Adeli K. Apolipoprotein B100 biogenesis: a complex array of intracellular mechanisms regulating folding, stability, and lipoprotein assembly. Biochem. Cell Biol. 2010;88:251–267. doi: 10.1139/o09-168. [DOI] [PubMed] [Google Scholar]
- 34.Olofsson SO, Boström P, Andersson L, Rutberg M, Perman J, Borén J. Lipid droplets as dynamic organelles connecting storage and efflux of lipids. Biochim. Biophys. Acta. 2009;1791:448–458. doi: 10.1016/j.bbalip.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 35.Merz A, Long G, Hiet M-SS, et al. Biochemical and morphological properties of hepatitis C virus particles and determination of their lipidome. J. Biol. Chem. 2011;286:3018–3032. doi: 10.1074/jbc.M110.175018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chang K-S, Jiang J, Cai Z, Luo G. Human apolipoprotein e is required for infectivity and production of hepatitis C virus in cell culture. J. Virol. 2007;81:13783–13793. doi: 10.1128/JVI.01091-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jiang J, Luo G. Apolipoprotein E but not B is required for the formation of infectious hepatitis C virus particles. J. Virol. 2009;83:12680–12691. doi: 10.1128/JVI.01476-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meunier J-C, Russell RS, Engle RE, Faulk KN, Purcell RH, Emerson SU. Apolipoprotein c1 association with hepatitis C virus. J. Virol. 2008;82:9647–9656. doi: 10.1128/JVI.00914-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Thomssen R, Bonk S, Thiele A. Density heterogeneities of hepatitis C virus in human sera due to the binding of beta-lipoproteins and immunoglobulins. Med. Microbiol. Immunol. 1993;182(6):329–34. doi: 10.1007/BF00191948. [DOI] [PubMed] [Google Scholar]
- 40.Trestard A, Bacq Y, Buzelay L, et al. Ultrastructural and physicochemical characterization of the hepatitis C virus recovered from the serum of an agammaglobulinemic patient. Arch. Virol. 1998;143:2241–2245. doi: 10.1007/s007050050455. [DOI] [PubMed] [Google Scholar]
- 41.Maillard P, Krawczynski K, Nitkiewicz J, et al. Nonenveloped nucleocapsids of hepatitis C virus in the serum of infected patients. J. Virol. 2001;75:8240–8250. doi: 10.1128/JVI.75.17.8240-8250.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nielsen SU, Bassendine MF, Burt AD, Martin C, Pumeechockchai W, Toms GL. Association between hepatitis C virus and very-low-density lipoprotein (VLDL)/LDL analyzed in iodixanol density gradients. J. Virol. 2006;80(5):2418–28. doi: 10.1128/JVI.80.5.2418-2428.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bukh J. A critical role for the chimpanzee model in the study of hepatitis C. Hepatology. 2004;39:1469–1475. doi: 10.1002/hep.20268. [DOI] [PubMed] [Google Scholar]
- 44.Lindenbach BD, Evans MJ, Syder AJ, et al. Complete replication of hepatitis C virus in cell culture. Science. 2005;309(5734):623–626. doi: 10.1126/science.1114016. [DOI] [PubMed] [Google Scholar]
- 45.Ndongo-Thiam N, Berthillon P, Errazuriz E, et al. Long-term propagation of serum hepatitis c virus (HCV) with production of enveloped HCV particles in human heparg hepatocytes. Hepatology. 2011;54:406–417. doi: 10.1002/hep.24386. [DOI] [PubMed] [Google Scholar]
- 46.Scholtes C, Ramière C, Rainteau D, et al. High plasma level of nucleocapsid-free envelope glycoprotein-positive lipoproteins in hepatitis C patients. Hepatology. 2012;56:39–48. doi: 10.1002/hep.25628. [DOI] [PubMed] [Google Scholar]
- 47.Bradley D, McCaustland K, Krawczynski K, Spelbring J, Humphrey C, Cook EHH. Hepatitis C virus: buoyant density of the factor VIII-derived isolate in sucrose. J. Med. Virol. 1991;34:206–208. doi: 10.1002/jmv.1890340315. [DOI] [PubMed] [Google Scholar]
- 48.Gastaminza P, Kapadia SB, Chisari FV. Differential biophysical properties of infectious intracellular and secreted hepatitis C virus particles. J. Virol. 2006;80:11074–11081. doi: 10.1128/JVI.01150-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lindenbach BD, Meuleman P, Ploss A, et al. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl Acad. Sci. USA. 2006;103:3805–3809. doi: 10.1073/pnas.0511218103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.André P, Komurian-Pradel F, Deforges S, et al. Characterization of low- and very-low-density hepatitis C virus RNA-containing particles. J. Virol. 2002;76:6919–6928. doi: 10.1128/JVI.76.14.6919-6928.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kono Y, Hayashida K, Tanaka H, Ishibashi H, Harada M. High-density lipoprotein binding rate differs greatly between genotypes 1b and 2a/2b of hepatitis C virus. J. Med. Virol. 2003;70:42–48. doi: 10.1002/jmv.10372. [DOI] [PubMed] [Google Scholar]
- 52.Diaz O, Delers F, Maynard M, et al. Preferential association of hepatitis C virus with apolipoprotein B48-containing lipoproteins. J. Gen. Virol. 2006;87:2983–2991. doi: 10.1099/vir.0.82033-0. Pt 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Podevin P, Carpentier A, Pne V, et al. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology. 2010;139 doi: 10.1053/j.gastro.2010.06.058. [DOI] [PubMed] [Google Scholar]
- 54.Dreux M, Boson B, Ricard-Blum S, et al. The exchangeable apolipoprotein ApoC-I promotes membrane fusion of hepatitis C virus. J. Biol. Chem. 2007;282(44):32357–32369. doi: 10.1074/jbc.M705358200. [DOI] [PubMed] [Google Scholar]
- 55.Miyanari Y, Atsuzawa K, Usuda N, et al. The lipid droplet is an important organelle for hepatitis C virus production. Nat. Cell Biol. 2007;9:1089–1097. doi: 10.1038/ncb1631. [DOI] [PubMed] [Google Scholar]
- 56.Gastaminza P, Dryden KA, Boyd B, et al. Ultrastructural and biophysical characterization of hepatitis C virus particles produced in cell culture. J. Virol. 2010;84:10999–11009. doi: 10.1128/JVI.00526-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Huang H, Sun F, Owen DM, et al. Hepatitis C virus production by human hepatocytes dependent on assembly and secretion of very low-density lipoproteins. Proc. Natl Acad. Sci. USA. 2007;104:5848–5853. doi: 10.1073/pnas.0700760104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nahmias Y, Goldwasser J, Casali M, et al. Apolipoprotein B-dependent hepatitis C virus secretion is inhibited by the grapefruit flavonoid naringenin. Hepatology. 2008;47:1437–1445. doi: 10.1002/hep.22197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lindenbach BD. Virion assembly and release. Curr. Top. Microbiol. Immunol. 2013;369:199–218. doi: 10.1007/978-3-642-27340-7_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sheridan DA, Bridge SH, Felmlee DJ, et al. Apolipoprotein-E and hepatitis C lipoviral particles in genotype 1 infection: evidence for an association with interferon sensitivity. J. Hepatol. 2012;57:32–38. doi: 10.1016/j.jhep.2012.02.017. [DOI] [PubMed] [Google Scholar]
- 61.Jiang J, Cun W, Wu X, Shi Q, Tang H, Luo G. Hepatitis C virus attachment mediated by apolipoprotein E binding to cell surface heparan sulfate. J. Virol. 2012;86:7256–7267. doi: 10.1128/JVI.07222-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jiang J, Wu X, Tang H, Luo G. Apolipoprotein E mediates attachment of clinical hepatitis C virus to hepatocytes by binding to cell surface heparan sulfate proteoglycan receptors. PLoS ONE. 2013;8 doi: 10.1371/journal.pone.0067982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Liu S, McCormick KD, Zhao W, Zhao T, Fan D, Wang T. Human apolipoprotein E peptides inhibit hepatitis C virus entry by blocking virus binding. Hepatology. 2012;56:484–491. doi: 10.1002/hep.25665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lefèvre M, Felmlee DJ, Parnot M, Baumert TF, Schuster C. Syndecan 4 is involved in mediating HCV entry through interaction with lipoviral particle-associated apolipoprotein E. PLoS ONE. 2014;9(4):e95550. doi: 10.1371/journal.pone.0095550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Maillard P, Huby T, Andréo U, Moreau M, Chapman J, Budkowska A. The interaction of natural hepatitis C virus with human scavenger receptor SR-BI/Cla1 is mediated by ApoB-containing lipoproteins. FASEB J. 2006;20(6):735–7. doi: 10.1096/fj.05-4728fje. [DOI] [PubMed] [Google Scholar]
- 66.Bankwitz D, Vieyres G, Hueging K, et al. Role of hypervariable region 1 for the interplay of hepatitis C virus with entry factors and lipoproteins. J. Virol. 2014;88(21):12644–55. doi: 10.1128/JVI.01145-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Van Eck M, Hoekstra M, Out R, et al. Scavenger receptor BI facilitates the metabolism of VLDL lipoproteins in vivo. J. Lipid Res. 2008;49:136–146. doi: 10.1194/jlr.M700355-JLR200. [DOI] [PubMed] [Google Scholar]
- 68.Hishiki T, Shimizu Y, Tobita R, et al. Infectivity of hepatitis C virus is influenced by association with apolipoprotein E isoforms. J. Virol. 2010;84:12048–12057. doi: 10.1128/JVI.01063-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Owen DM, Huang H, Ye J, Gale M. Apolipoprotein E on hepatitis C virion facilitates infection through interaction with low-density lipoprotein receptor. Virology. 2009;394(1):99–108. doi: 10.1016/j.virol.2009.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Albecka A, Belouzard S, Op de Beeck A, et al. Role of low-density lipoprotein receptor in the hepatitis C virus life cycle. Hepatology. 2012;55:998–1007. doi: 10.1002/hep.25501. [DOI] [PubMed] [Google Scholar]
- 71.Pileri P. Binding of hepatitis C virus to CD81. Science. 1998;282(5390):938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 72.Bartosch B, Dubuisson J, Cosset F-L. Infectious hepatitis C virus pseudo-particles containing functional E1-E2 envelope protein complexes. J. Exp. Med. 2003;197(5):633–642. doi: 10.1084/jem.20021756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Cormier EG, Tsamis F, Kajumo F, Durso RJ, Gardner JP, Dragic T. CD81 is an entry coreceptor for hepatitis C virus. Proc. Natl Acad. Sci. USA. 2004;101(19):7270–4. doi: 10.1073/pnas.0402253101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang J, Randall G, Higginbottom A, Rice CM, Mckeating JA, Monk P. CD81 is required for hepatitis C virus glycoprotein-mediated viral infection. J. Virol. 2004;78(3):1448–1455. doi: 10.1128/JVI.78.3.1448-1455.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hsu M, Zhang J, Flint M, et al. Hepatitis C virus glycoproteins mediate pH-dependent cell entry of pseudotyped retroviral particles. Proc. Natl Acad. Sci. USA. 2003;100(12):7271–7276. doi: 10.1073/pnas.0832180100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhong J, Gastaminza P, Cheng G, et al. Robust hepatitis C virus infection in vitro. Proc. Natl Acad. Sci. USA. 2005;102:9294–9299. doi: 10.1073/pnas.0503596102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wakita T, Pietschmann T, Kato T, et al. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 2005;11:791–796. doi: 10.1038/nm1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cai Z, Zhang C, Chang K-S, et al. Robust production of infectious hepatitis C virus (HCV) from stably HCV cDNA-transfected human hepatoma cells. J. Virol. 2005;79:13963–13973. doi: 10.1128/JVI.79.22.13963-13973.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Akazawa D, Date T, Morikawa K, et al. CD81 expression is important for the permissiveness of Huh7 cell clones for heterogeneous hepatitis C virus infection. J. Virol. 2007;81:5036–5045. doi: 10.1128/JVI.01573-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ploss A, Evans MJ, Gaysinskaya V a, et al. Human occludin is a hepatitis C virus entry factor required for infection of mouse cells. Nature. 2009;457(7231):882–886. doi: 10.1038/nature07684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Meuleman P, Hesselgesser J, Paulson M, et al. Anti-CD81 antibodies can prevent a hepatitis C virus infection in vivo. Hepatology. 2008;48(6):1761–1768. doi: 10.1002/hep.22547. [DOI] [PubMed] [Google Scholar]
- 82.Dorner M, Horwitz JA, Robbins JB, et al. A genetically humanized mouse model for hepatitis C virus infection. Nature. 2011;474:208–211. doi: 10.1038/nature10168. • Reports the development of the first immunocompetent nontransplant murine model for hepatitis C virus entry, providing a system for in vivo studies. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Levy S, Nguyen VQ, Andria ML, Takahashi S. Structure and membrane topology of TAPA-1. J Biol Chem. 1991;266:14597–14602. [PubMed] [Google Scholar]
- 84.Seigneuret M. Complete predicted three-dimensional structure of the facilitator transmembrane protein and hepatitis C virus receptor CD81: conserved and variable structural domains in the tetraspanin superfamily. Biophys. J. 2006;90(1):212–227. doi: 10.1529/biophysj.105.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Krey T, D’Alayer J, Kikuti CM, et al. The disulfide bonds in glycoprotein E2 of hepatitis C virus reveal the tertiary organization of the molecule. PLoS Pathog. 2010;6(2):e1000762. doi: 10.1371/journal.ppat.1000762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wahid A, Helle F, Descamps V, Duverlie G, Penin F, Dubuisson J. Disulfide bonds in hepatitis C virus glycoprotein E1 control the assembly and entry functions of E2 glycoprotein. J. Virol. 2013;87(3):1605–17. doi: 10.1128/JVI.02659-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Scarselli E, Ansuini H, Cerino R, et al. The human scavenger receptor class B type I is a novel candidate receptor for the hepatitis C virus. EMBO J. 2002;21(19):5017–5025. doi: 10.1093/emboj/cdf529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Voisset C, Callens N, Blanchard E, Op De Beeck A, Dubuisson J, Vu-Dac N. High density lipoproteins facilitate hepatitis C virus entry through the scavenger receptor class B type I. J. Biol. Chem. 2005;280(9):7793–9. doi: 10.1074/jbc.M411600200. [DOI] [PubMed] [Google Scholar]
- 89.Lavillette D, Tarr AW, Voisset C, et al. Characterization of host-range and cell entry properties of the major genotypes and subtypes of hepatitis C virus. Hepatology. 2005;41(2):265–74. doi: 10.1002/hep.20542. [DOI] [PubMed] [Google Scholar]
- 90.Dreux M, Pietschmann T, Granier C, et al. High density lipoprotein inhibits hepatitis C virus-neutralizing antibodies by stimulating cell entry via activation of the scavenger receptor BI. J. Biol. Chem. 2006;281:18285–18295. doi: 10.1074/jbc.M602706200. [DOI] [PubMed] [Google Scholar]
- 91.Kapadia SB, Barth H, Baumert T, McKeating JA, Chisari FV. Initiation of hepatitis C virus infection is dependent on cholesterol and cooperativity between CD81 and scavenger receptor B type I. J. Virol. 2007;81(1):374–83. doi: 10.1128/JVI.01134-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Zeisel MB, Koutsoudakis G, Schnober EK, et al. Scavenger receptor class B type I is a key host factor for hepatitis C virus infection required for an entry step closely linked to CD81. Hepatology. 2007;46(6):1722–31. doi: 10.1002/hep.21994. [DOI] [PubMed] [Google Scholar]
- 93.Thi VLD, Granier C, Zeisel MB, et al. Characterization of hepatitis C virus particle subpopulations reveals multiple usage of the scavenger receptor BI for entry steps. J. Biol. Chem. 2012;287:31242–31257. doi: 10.1074/jbc.M112.365924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pöhlmann S, Zhang J, Baribaud F, et al. Hepatitis C virus glycoproteins interact with DC-SIGN and DC-SIGNR. J. Virol. 2003;77:4070–4080. doi: 10.1128/JVI.77.7.4070-4080.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lai WK, Sun PJ, Zhang J, et al. Expression of DC-SIGN and DC-SIGNR on human sinusoidal endothelium: a role for capturing hepatitis C virus particles. Am. J. Pathol. 2006;169:200–208. doi: 10.2353/ajpath.2006.051191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Koutsoudakis G, Kaul A, Steinmann E, et al. Characterization of the early steps of hepatitis C virus infection by using luciferase reporter viruses. J. Virol. 2006;80:5308–5320. doi: 10.1128/JVI.02460-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Barth H, Schäfer C, Adah MI, et al. Cellular binding of hepatitis C virus envelope glycoprotein E2 requires cell surface heparan sulfate. J. Biol. Chem. 2003;278:41003–41012. doi: 10.1074/jbc.M302267200. [DOI] [PubMed] [Google Scholar]
- 98.Shi Q, Jiang J, Luo G. Syndecan-1 serves as the major receptor for attachment of hepatitis C virus to the surfaces of hepatocytes. J. Virol. 2013;87(12):6866–6875. doi: 10.1128/JVI.03475-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Gong Q, Cheng M, Chen H, et al. Phospholipid scramblase 1 mediates hepatitis C virus entry into host cells. FEBS Lett. 2011;585:2647–2652. doi: 10.1016/j.febslet.2011.07.019. [DOI] [PubMed] [Google Scholar]
- 100.Mazumdar B, Banerjee A, Meyer K, Ray R. Hepatitis C virus E1 envelope glycoprotein interacts with apolipoproteins in facilitating entry into hepatocytes. Hepatology. 2011;54:1149–1156. doi: 10.1002/hep.24523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yi M, Kaneko S, Yu DYY, Murakami S. Hepatitis C virus envelope proteins bind lactoferrin. J. Virol. 1997;71:5997–6002. doi: 10.1128/jvi.71.8.5997-6002.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Evans MJ, von Hahn T, Tscherne DM, et al. Claudin-1 is a hepatitis C virus co-receptor required for a late step in entry. Nature. 2007;446(7137):801–805. doi: 10.1038/nature05654. [DOI] [PubMed] [Google Scholar]
- 103.Reynolds GM, Harris HJ, Jennings A, et al. Hepatitis C virus receptor expression in normal and diseased liver tissue. Hepatology. 2008;47(2):418–27. doi: 10.1002/hep.22028. [DOI] [PubMed] [Google Scholar]
- 104.Harris HJ, Farquhar MJ, Mee CJ, et al. CD81 and claudin 1 coreceptor association: role in hepatitis C virus entry. J. Virol. 2008;82:5007–5020. doi: 10.1128/JVI.02286-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Harris HJ, Davis C, Mullins JGL, et al. Claudin association with CD81 defines hepatitis C virus entry. J. Biol. Chem. 2010;285(27):21092–102. doi: 10.1074/jbc.M110.104836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Farquhar MJ, Harris HJ, Diskar M, et al. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 2008;82:8797–8811. doi: 10.1128/JVI.00592-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Douam F, Dao Thi VL, Maurin G, et al. Critical interaction between E1 and E2 glycoproteins determines binding and fusion properties of hepatitis C virus during cell entry. Hepatology. 2014;59(3):776–788. doi: 10.1002/hep.26733. [DOI] [PubMed] [Google Scholar]
- 108.Zheng A, Yuan F, Li Y, et al. Claudin-6 and claudin-9 function as additional coreceptors for hepatitis C virus. J. Virol. 2007;81(22):12465–1271. doi: 10.1128/JVI.01457-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Meertens L, Bertaux C, Cukierman L, et al. The tight junction proteins claudin-1, -6, and -9 are entry cofactors for hepatitis C virus. J. Virol. 2008;82:3555–3560. doi: 10.1128/JVI.01977-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Haid S, Grethe C, Dill MT, Heim M, Kaderali L, Pietschmann T. Isolate-dependent use of claudins for cell entry by hepatitis C virus. Hepatology. 2014;59(1):24–34. doi: 10.1002/hep.26567. [DOI] [PubMed] [Google Scholar]
- 111.Fofana I, Zona L, Thumann C, et al. Functional analysis of claudin-6 and claudin-9 as entry factors for hepatitis C virus infection of human hepatocytes by using monoclonal antibodies. J. Virol. 2013;87:10405–10410. doi: 10.1128/JVI.01691-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Liu S, Yang W, Shen L, Turner JR, Coyne CB, Wang T. Tight junction proteins claudin-1 and occludin control hepatitis C virus entry and are downregulated during infection to prevent superinfection. J. Virol. 2009;83:2011–2014. doi: 10.1128/JVI.01888-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dorner M, Horwitz J a, Donovan BM, et al. Completion of the entire hepatitis C virus life cycle in genetically humanized mice. Nature. 2013;501:237–241. doi: 10.1038/nature12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Li Q, Zhang Y, Chiu S, et al. Integrative functional genomics of hepatitis C virus infection identifies host dependencies in complete viral replication cycle. PLoS Pathog. 2014;10:e1004163. doi: 10.1371/journal.ppat.1004163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Lupberger J, Zeisel MB, Xiao F, et al. EGFR and EphA2 are host factors for hepatitis C virus entry and possible targets for antiviral therapy. Nat. Med. 2011;17:589–595. doi: 10.1038/nm.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Coller KE, Berger KL, Heaton NS, Cooper JD, Yoon R, Randall G. RNA interference and single particle tracking analysis of hepatitis C virus endocytosis. PLoS Pathog. 2009;5:e1000702. doi: 10.1371/journal.ppat.1000702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Diao J, Pantua H, Ngu H, et al. Hepatitis C virus (HCV) induces epidermal growth factor receptor (EGFR) activation via CD81 binding for viral internalization and entry. J. Virol. 2012;86(8):4305–4316. doi: 10.1128/JVI.00750-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zona L, Lupberger J, Sidahmed-Adrar N, et al. HRas signal transduction promotes hepatitis C virus cell entry by triggering assembly of the host tetraspanin receptor complex. Cell Host Microbe. 2013;13:302–313. doi: 10.1016/j.chom.2013.02.006. [DOI] [PubMed] [Google Scholar]
- 119.Aizaki H, Morikawa K, Fukasawa M, et al. Critical role of virion-associated cholesterol and sphingolipid in hepatitis C virus infection. J. Virol. 2008;82:5715–5724. doi: 10.1128/JVI.02530-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sainz B, Barretto N, Martin DN, et al. Identification of the Niemann-Pick C1–like 1 cholesterol absorption receptor as a new hepatitis C virus entry factor. Nat. Med. 2012;18:281–285. doi: 10.1038/nm.2581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Altmann SW, Davis HR, Zhu L-J, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 122.Ye J. Reliance of host cholesterol metabolic pathways for the life cycle of hepatitis C virus. PLoS Pathog. 2007;3(8):e108. doi: 10.1371/journal.ppat.0030108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Côté M, Misasi J, Ren T, et al. Small molecule inhibitors reveal Niemann-Pick C1 is essential for ebola virus infection. Nature. 2011;477:344–348. doi: 10.1038/nature10380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Carette JEE, Raaben M, Wong ACC, et al. Ebola virus entry requires the cholesterol transporter Niemann-Pick C1. Nature. 2011;477:340–343. doi: 10.1038/nature10348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Wu X, Robotham JM, Lee E, et al. Productive hepatitis C virus infection of stem cell-derived hepatocytes reveals a critical transition to viral permissiveness during differentiation. PLoS Pathog. 2012;8:e1002617. doi: 10.1371/journal.ppat.1002617. • Demonstrates the feasibility of using stem cell differentiation to identify the transition to viral permissiveness, which in turn enables discovery of host factors for viral infection. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wu X, Lee EM, Hammack C, et al. Cell death-inducing DFFA-like effector b is required for hepatitis C virus entry into hepatocytes. J. Virol. 2014;88(15):8433–44. doi: 10.1128/JVI.00081-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Kim S, Ishida H, Yamane D, et al. Contrasting roles of mitogen-activated protein kinases in cellular entry and replication of hepatitis C virus: MKNK1 facilitates cell entry. J. Virol. 2013;87(8):4214–4224. doi: 10.1128/JVI.00954-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Monazahian M, Böhme I, Bonk S, et al. Low density lipoprotein receptor as a candidate receptor for hepatitis C virus. J. Med. Virol. 1999;57:223–229. doi: 10.1002/(sici)1096-9071(199903)57:3<223::aid-jmv2>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 129.Martin DN, Uprichard SL. Identification of transferrin receptor 1 as a hepatitis C virus entry factor. Proc. Natl Acad. Sci. USA. 2013;110(26):10777–10782. doi: 10.1073/pnas.1301764110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Easter DW, Wade JB, Boyer JL. Structural integrity of hepatocyte tight junctions. J. Cell Biol. 1983;96(3):745–749. doi: 10.1083/jcb.96.3.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Harris HJ, Wilson GK, Hübscher SG, McKeating JA. Heterogeneous claudin-1 expression in human liver. Hepatology. 2013;57(2):854–5. doi: 10.1002/hep.25910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Wilson GK, Brimacombe CL, Rowe IA, et al. A dual role for hypoxia inducible factor-1α in the hepatitis C virus lifecycle and hepatoma migration. J. Hepatol. 2012;56:803–809. doi: 10.1016/j.jhep.2011.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Mee CJ, Farquhar MJ, Harris HJ, et al. Hepatitis C virus infection reduces hepatocellular polarity in a vascular endothelial growth factor-dependent manner. Gastroenterology. 2010;138:1134–1142. doi: 10.1053/j.gastro.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Benedicto I, Molina-Jiménez F, Barreiro O, et al. Hepatitis C virus envelope components alter localization of hepatocyte tight junction-Associated proteins and promote occludin retention in the endoplasmic reticulum. Hepatology. 2008;48(4):1044–1053. doi: 10.1002/hep.22465. [DOI] [PubMed] [Google Scholar]
- 135.Chiu J-H, Hu C-P, Lui W-Y, Lo SJ, Chang C. The formation of bile canaliculi in human hepatoma cell lines. Hepatology. 1990;11(5):834–842. doi: 10.1002/hep.1840110519. [DOI] [PubMed] [Google Scholar]
- 136.Traber MG, Kayden HJ, Rindler MJ. Polarized secretion of newly synthesized lipoproteins by the Caco-2 human intestinal cell line. J. Lipid Res. 1987;28:1350–1363. [PubMed] [Google Scholar]
- 137.Ratcliffe DR, Iqbal J, Hussain MM, Cramer EB. Fibrillar collagen type I stimulation of apolipoprotein B secretion in Caco-2 cells is mediated by beta1 integrin. Biochim. Biophys. Acta. 2009;1791:1144–1154. doi: 10.1016/j.bbalip.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Keck Z-Y, Xia J, Cai Z, et al. Immunogenic and functional organization of hepatitis C virus (HCV) glycoprotein E2 on infectious HCV virions. J. Virol. 2007;81:1043–1047. doi: 10.1128/JVI.01710-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Stamataki Z, Coates S, Evans MJ, et al. Hepatitis C virus envelope glycoprotein immunization of rodents elicits cross-reactive neutralizing antibodies. Vaccine. 2007;25:7773–7784. doi: 10.1016/j.vaccine.2007.08.053. [DOI] [PubMed] [Google Scholar]
- 140.Owsianka AM, Tarr AW, Keck Z-Y, et al. Broadly neutralizing human monoclonal antibodies to the hepatitis C virus E2 glycoprotein. J. Gen. Virol. 2008;89:653–659. doi: 10.1099/vir.0.83386-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Meunier J-C, Gottwein JM, Houghton M, et al. Vaccine-induced cross-genotype reactive neutralizing antibodies against hepatitis C virus. J. Infect. Dis. 2011;204:1186–1190. doi: 10.1093/infdis/jir511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Mee CJ, Harris HJ, Farquhar MJ, et al. Polarization restricts hepatitis C virus entry into HepG2 hepatoma cells. J. Virol. 2009;83:6211–6221. doi: 10.1128/JVI.00246-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Narbus CM, Israelow B, Sourisseau M, et al. HepG2 cells expressing microRNA miR-122 support the entire hepatitis C virus life cycle. J. Virol. 2011;85:12087–12092. doi: 10.1128/JVI.05843-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sainz B, TenCate V, Uprichard SL. Three-dimensional Huh7 cell culture system for the study of hepatitis C virus infection. Virol. J. 2009;6:103. doi: 10.1186/1743-422X-6-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Molina-Jimenez F, Benedicto I, Dao Thi VL, et al. Matrigel-embedded 3D culture of Huh-7 cells as a hepatocyte-like polarized system to study hepatitis C virus cycle. Virology. 2012;425:31–39. doi: 10.1016/j.virol.2011.12.021. [DOI] [PubMed] [Google Scholar]
- 146.Carloni G, Iacovacci S, Sargiacomo M, et al. Susceptibility of human liver cell cultures to hepatitis C virus infection. Arch. Virol. Suppl. 1993;8:31–39. doi: 10.1007/978-3-7091-9312-9_4. [DOI] [PubMed] [Google Scholar]
- 147.Fournier C, Sureau C, Coste J, et al. In vitro infection of adult normal human hepatocytes in primary culture by hepatitis C virus. J. Gen. Virol. 1998;79:2367–2374. doi: 10.1099/0022-1317-79-10-2367. [DOI] [PubMed] [Google Scholar]
- 148.Rumin S, Berthillon P, Tanaka E, et al. Dynamic analysis of hepatitis C virus replication and quasispecies selection in long-term cultures of adult human hepatocytes infected in vitro. J. Gen. Virol. 1999;80:3007–3018. doi: 10.1099/0022-1317-80-11-3007. [DOI] [PubMed] [Google Scholar]
- 149.Lázaro CA, Chang M, Tang W, et al. Hepatitis C virus replication in transfected and serum-infected cultured human fetal hepatocytes. Am. J. Pathol. 2007;170:478–489. doi: 10.2353/ajpath.2007.060789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Molina S, Castet V, Pichard-Garcia L, et al. Serum-derived hepatitis C virus infection of primary human hepatocytes is tetraspanin CD81 dependent. J. Virol. 2008;82:569–574. doi: 10.1128/JVI.01443-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Ploss A, Khetani SR, Jones CT, et al. Persistent hepatitis C virus infection in microscale primary human hepatocyte cultures. Proc. Natl Acad. Sci. USA. 2010;107:3141–3145. doi: 10.1073/pnas.0915130107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Gondeau C, Briolotti P, Razafy F, et al. In vitro infection of primary human hepatocytes by HCV-positive sera: insights on a highly relevant model. Gut. 2014;63:1490–500. doi: 10.1136/gutjnl-2013-304623. [DOI] [PubMed] [Google Scholar]
- 153.Schwartz RE, Trehan K, Andrus L, et al. Modeling hepatitis C virus infection using human induced pluripotent stem cells. Proc. Natl Acad. Sci. USA. 2012;109:2544–8. doi: 10.1073/pnas.1121400109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Roelandt P, Obeid S, Paeshuyse J, et al. Human pluripotent stem cell-derived hepatocytes support complete replication of hepatitis C virus. J. Hepatol. 2012;57:246–251. doi: 10.1016/j.jhep.2012.03.030. [DOI] [PubMed] [Google Scholar]
- 155.Codran A, Royer C, Jaeck D, et al. Entry of hepatitis C virus pseudotypes into primary human hepatocytes by clathrin-dependent endocytosis. J. Gen. Virol. 2006;87:2583–2593. doi: 10.1099/vir.0.81710-0. Pt 9. [DOI] [PubMed] [Google Scholar]
- 156.Blanchard E, Belouzard S, Goueslain L, et al. Hepatitis C virus entry depends on clathrin-mediated endocytosis. J. Virol. 2006;80(14):6964–6972. doi: 10.1128/JVI.00024-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Neill JD. Molecular biology of bovine viral diarrhea virus. Biologicals. 2013;41(1):2–7. doi: 10.1016/j.biologicals.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 158.Sánchez-San Martín C, Liu CY, Kielian M. Dealing with low pH: entry and exit of alphaviruses and flaviviruses. Trends Microbiol. 2009;17(11):514–521. doi: 10.1016/j.tim.2009.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Garry RF, Dash S. Proteomics computational analyses suggest that hepatitis C virus E1 and pestivirus E2 envelope glycoproteins are truncated class II fusion proteins. Virology. 2003;307(2):255–265. doi: 10.1016/s0042-6822(02)00065-x. [DOI] [PubMed] [Google Scholar]
- 160.Vaney M-C, Rey FA. Class II enveloped viruses. Cell Microbiol. 2011;13(10):1451–9. doi: 10.1111/j.1462-5822.2011.01653.x. [DOI] [PubMed] [Google Scholar]
- 161.Rey FA, Heinz FX, Mandl C, Kunz C, Harrison SC. The envelope glycoprotein from tick-borne encephalitis virus at 2 A resolution. Nature. 1995;375:291–298. doi: 10.1038/375291a0. [DOI] [PubMed] [Google Scholar]
- 162.Modis Y, Ogata S, Clements D, Harrison SC. A ligand-binding pocket in the dengue virus envelope glycoprotein. Proc. Natl Acad. Sci. USA. 2003;100:6986–6991. doi: 10.1073/pnas.0832193100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Nybakken GE, Nelson CA, Chen BR, Diamond MS, Fremont DH. Crystal structure of the West Nile virus envelope glycoprotein. J. Virol. 2006;80:11467–11474. doi: 10.1128/JVI.01125-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Li L, Lok S-M, Yu I-M, et al. The flavivirus precursor membrane-envelope protein complex: structure and maturation. Science. 2008;319:1830–1834. doi: 10.1126/science.1153263. [DOI] [PubMed] [Google Scholar]
- 165.Lescar J, Roussel A, Wien MW, et al. The fusion glycoprotein shell of Semliki Forest virus: an icosahedral assembly primed for fusogenic activation at endosomal pH. Cell. 2001;105:137–148. doi: 10.1016/s0092-8674(01)00303-8. [DOI] [PubMed] [Google Scholar]
- 166.Roussel A, Lescar J, Vaney MC, Wengler G, Wengler G, Rey FA. Structure and interactions at the viral surface of the envelope protein E1 of semliki forest virus. Structure. 2006;14:75–86. doi: 10.1016/j.str.2005.09.014. [DOI] [PubMed] [Google Scholar]
- 167.Li L, Jose J, Xiang Y, Kuhn RJ, Rossmann MG. Structural changes of envelope proteins during alphavirus fusion. Nature. 2010;468:705–708. doi: 10.1038/nature09546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Voss JE, Vaney M-C, Duquerroy S, et al. Glycoprotein organization of chikungunya virus particles revealed by x-ray crystallography. Nature. 2010;468:709–712. doi: 10.1038/nature09555. [DOI] [PubMed] [Google Scholar]
- 169.El Omari K, Iourin O, Harlos K, Grimes JM, Stuart DI. Structure of a pestivirus envelope glycoprotein E2 clarifies its role in cell entry. Cell Rep. 2013;3(1):30–5. doi: 10.1016/j.celrep.2012.12.001. • This paper presents the crystal structure of the BVDV-1 E2 glycoprotein, the structure of which suggests a mechanism of fusion that is distinct from that of flavivirus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Li Y, Wang J, Kanai R, Modis Y. Crystal structure of glycoprotein E2 from bovine viral diarrhea virus. Proc. Natl Acad. Sci. USA. 2013;110:6805–10. doi: 10.1073/pnas.1300524110. • Also provides key insights into BVDV-1 E2 3D architecture and how this glycoprotien dramatically differs from the fusion proteins of flaviviruses. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Iqbal M, McCauley JWW. Identification of the glycosaminoglycan-binding site on the glycoprotein Erns of bovine viral diarrhoea virus by site-directed mutagenesis. J. Gen. Virol. 2002;83:2153–2159. doi: 10.1099/0022-1317-83-9-2153. [DOI] [PubMed] [Google Scholar]
- 172.Magkouras I, Mätzener P, Rümenapf T, Peterhans E, Schweizer M. RNase-dependent inhibition of extracellular, but not intracellular, dsRNA-induced interferon synthesis by Erns of pestiviruses. J. Gen. Virol. 2008;89:2501–2506. doi: 10.1099/vir.0.2008/003749-0. [DOI] [PubMed] [Google Scholar]
- 173.Mätzener P, Magkouras I, Rümenapf T, Peterhans E, Schweizer M. The viral RNase Erns prevents IFN type-I triggering by pestiviral single- and double-stranded RNAs. Virus Res. 2009;140:15–23. doi: 10.1016/j.virusres.2008.10.015. [DOI] [PubMed] [Google Scholar]
- 174.Flint M, Thomas JM, Maidens CM, et al. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J. Virol. 1999;73(8):6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Drummer HE, Boo I, Poumbourios P. Mutagenesis of a conserved fusion peptide-like motif and membrane-proximal heptad-repeat region of hepatitis C virus glycoprotein E1. J. Gen. Virol. 2007;88:1144–1148. doi: 10.1099/vir.0.82567-0. [DOI] [PubMed] [Google Scholar]
- 176.Li H-F, Huang C-H, Ai L-S, Chuang C-K, Chen SSL. Mutagenesis of the fusion peptide-like domain of hepatitis C virus E1 glycoprotein: involvement in cell fusion and virus entry. J. Biomed. Sci. 2009;16(1):89. doi: 10.1186/1423-0127-16-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Lavillette D, Pécheur E-I, Donot P, et al. Characterization of fusion determinants points to the involvement of three discrete regions of both E1 and E2 glycoproteins in the membrane fusion process of hepatitis C virus. J. Virol. 2007;81:8752–8765. doi: 10.1128/JVI.02642-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Op De Beeck A, Voisset C, Bartosch B, et al. Characterization of functional hepatitis C virus envelope glycoproteins. J Virol. 2004;78:2994–3002. doi: 10.1128/JVI.78.6.2994-3002.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]