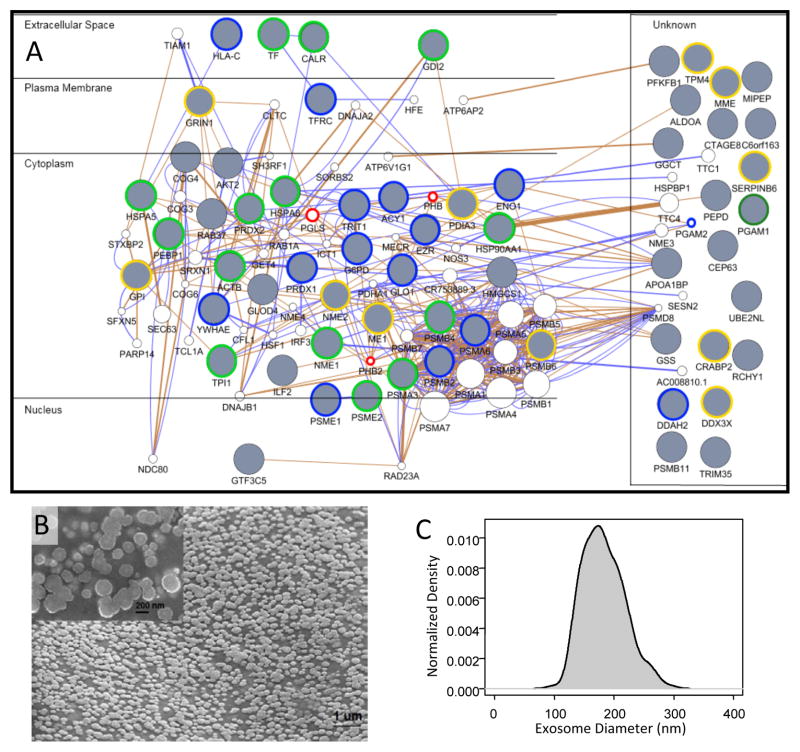
Figure 3. Protein-protein interaction networks suggest that the majority of proteins identified in both SKBR3 and BT474-conditioned media are derived from exosomes.
Protein-protein interaction networks were constructed in GeneMania using the union set of all identified proteins in SKBR3 and BT474 secretomes (Panel A), using just the SKBR3 secretome (Supplemental Figure S1), and using just the BT474 secretome (Supplemental Figure S2). The proteins were arranged based upon similarity in subcellular location. Gray circles indicate identified proteins while white circles indicate proteins inferred based upon known interactions with identified proteins. The blue edges indicate experimentally observed protein-protein interactions in Homo Sapiens, while brown edges are predicted protein-protein interactions based on homology with other species. Encircled nodes are found in the Exocarta database, which suggests that they are associated with exosomes. Nodes encircled in blue are from SKBR3, in yellow from BT474, and in green are common between SKBR4 and BT474. The nodes encircled in red are inferred only in the amalgamated network and are associated with exosomes. Scanning electron microscopy was used to confirm that the secretome contained membrane vesicles (Panel B - SKBR3-conditioned media). The size and distribution of membrane vesicles were consistent with exosomes (Panel C). SEM results representative of a least two replicates.