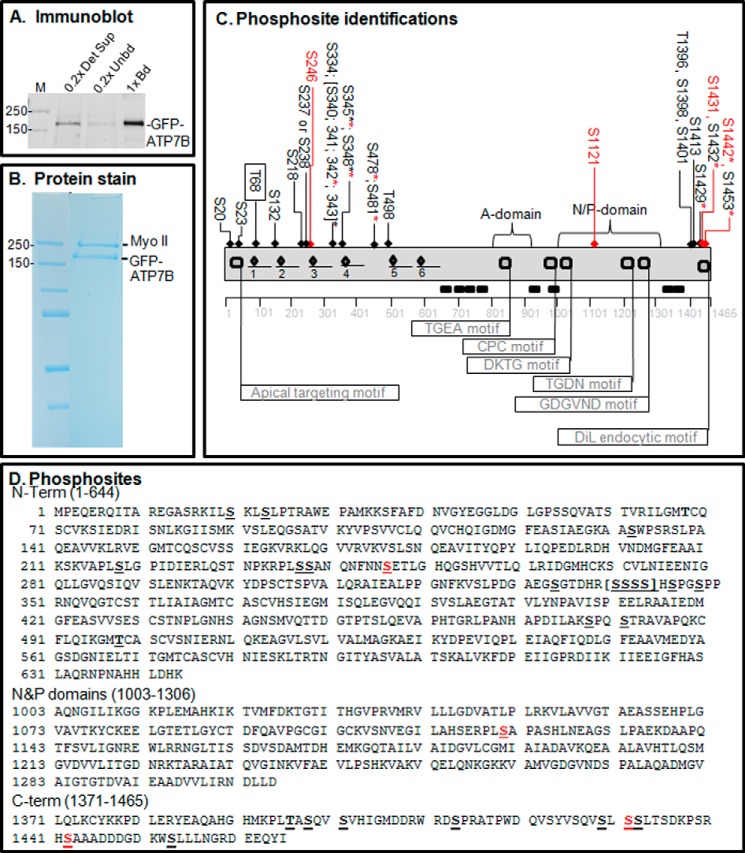
FIGURE 2.
Proteomics analysis of copper-dependent and copper-independent phosphorylation. A, YS cells expressing GFP-WTATP7B were extracted in detergent, and the ATP7B was immunoprecipitated. The gel shown is an immunoblot of the starting extract (0.2× Det Sup; left), depleted supernatant (0.2× Unbd; middle) and immunoprecipitated ATP7B protein (1× bound (Bd); right); molecular mass markers are indicated in kDa (far left). B, GFP-WTATP7B was immunoprecipitated, separated by SDS-PAGE, and then stained in-gel with Simply Blue. Gel bands of five independent samples for each treatment were used for MS analysis, and three and four independent MS analyses were performed for ±copper-treated cells, respectively. C, schematic depiction of phosphosites identified by proteomics in the context of the ATP7B domain structure (flags show the locations of characterized motifs). An underline shows the location of the six metal binding domains, and diamonds indicate the CXXC motifs that bind copper. All of the sites identified with >95% confidence are indicated across the top of the diagram; those highlighted in red were found only in the high copper condition. The boxed site near the N terminus was found only in the chelation condition. Asterisks indicate residues found as part of a diphosphopeptide or triphosphopeptide. See the supplemental data for documentation of the mass spectroscopy data; PEAKS analysis showed that ATP7B sequence coverage was 84%. D, phosphosite identifications mapped onto the ATP7B primary sequence. Red Ser residues were hyperphosphorylated in the presence of copper. DiL, dileucine.