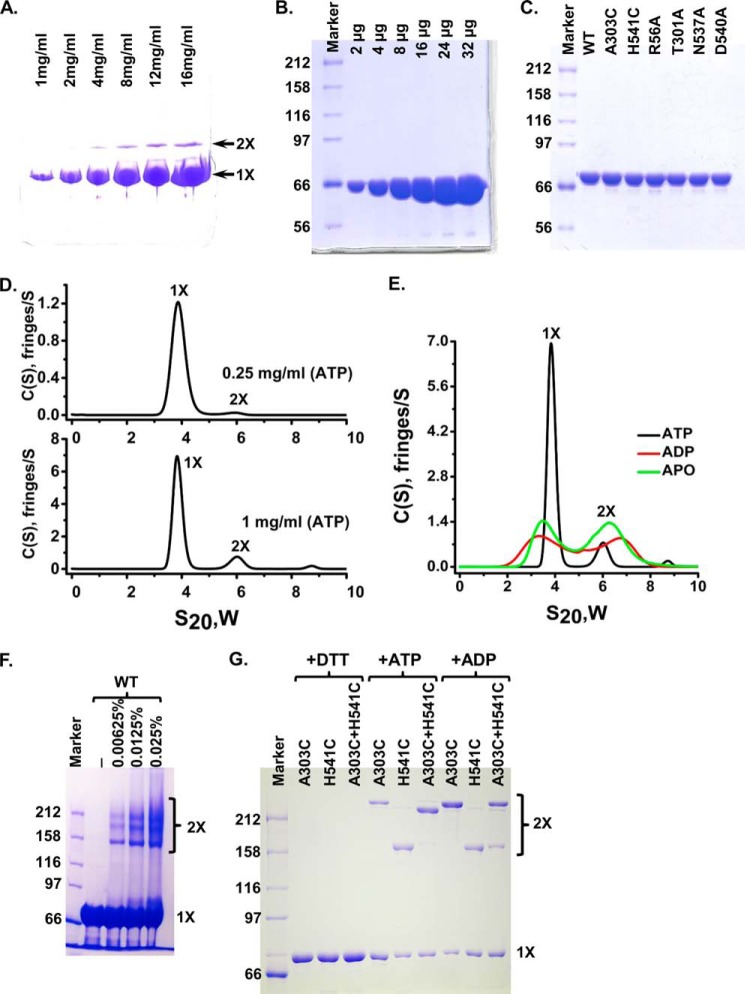
FIGURE 2.
DnaK forms specific dimer in solution in the presence of ATP. A, native PAGE analysis of DnaK-T199A/L′3,4. 2 μl of DnaK-T199A/L′3,4 with indicated concentrations were loaded onto an 8–25% gradient PhastGel (GE Healthcare). The positions of the monomer (1×) and dimer (2×) are indicated by arrows. B, DnaK-T199A/L′3,4 was purified to high purity. Equal amounts of DnaK-T199A/L′3,4 for each lane from A were loaded onto SDS-PAGE. There are no apparent contaminants in the purified DnaK-T199A/L′3,4 protein. C, mutant DnaK proteins were purified to a similar purity as that of the WT DnaK. Each mutant protein was loaded and separated on SDS-polyacrylamide gel. WT DnaK protein purified to high purity was used for comparison. D, AUC analysis of the WT DnaK protein in the presence of ATP. Sedimentation velocity experiments were carried out with WT DnaK protein at 0.25 (top panel) and 1 mg/ml (bottom panel). The positions of the monomer and dimer are labeled as 1× and 2×, respectively. E, AUC analysis of the WT DnaK protein in the presence of different nucleotides. Sedimentation velocity experiments were performed on WT DnaK protein at 1 mg/ml in the presence of ATP, ADP, or in the absence of nucleotide (apo-form). F, glutaraldehyde cross-linking. WT DnaK protein was treated with 0.00625, 0.0125, and 0.025% glutaraldehyde and separated on SDS-PAGE. The concentrations of glutaraldehyde were labeled on the top of the gel. The positions of the monomer (1×) and dimer (2×) are labeled on the right. G, disulfide bond formation between A303C and H541C. A303C, DnaK-C15A/A303C; H541C, DnaK-C15A/H541C; A303C+H541C, DnaK-C15A/A303C and DnaK-C15A/H541C were mixed in 1:1 ratio. Oxidation with copper-phenanthroline was carried out in the presence of ATP (+ATP) or ADP (+ADP). Samples in the presence of ATP were treated with DTT as loading controls (+DTT). The positions of the monomer (1×) and dimer (2×) are labeled on the right.